Pharmaceutical Sciences. 2025;31(3):237-250.
doi: 10.34172/PS.025.42140
Research Article
Development and Characterization of Liposomal Phytoformulation of Piper longum L. for Melanoma Therapy
Himani Himani Data curation, Methodology, Validation, Writing – review & editing, 1 
Debadatta Mohapatra Conceptualization, Data curation, Methodology, Software, Validation, Writing – original draft, 1 
Singh Shreya Formal analysis, Methodology, Validation, 1, 2 
Satyajeet Biswal Data curation, Formal analysis, Methodology, Software, Validation, 3 
Gaurav Gopal Naik Formal analysis, 1 
Soki Daeme Malang Formal analysis, Visualization, Writing – review & editing, 1 
Pooja Kathait Formal analysis, Visualization, Writing – review & editing, 1 
Pradeep Kumar Patel Formal analysis, Visualization, Writing – review & editing, 1 
Shambhavi Shambhavi Formal analysis, Visualization, Writing – review & editing, 1 
Prakash Ch. Senapati Investigation, Project administration, Supervision, 4 
Pratap Chandra Acharya Investigation, Project administration, Resources, Supervision, 3 
Alakh N Sahu Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, 1, * 
Author information:
1Phytomedicine Research Laboratory, Department of Pharmaceutical Engineering & Technology, Indian Institute of Technology (BHU), Varanasi-221005, India
2Department of Pharmacology, SBS College of Pharmacy, Malwan, Fatehpur-212664, Indi
3Drug Metabolomics Laboratory, Department of Pharmacy, Tripura University (A Central University), Suryamaninagar-799022, Tripura, India
4Department of Pharmaceutics, IMT Pharmacy College, Gopalpur, Puri-752004, Odisha, Indi
Abstract
Background:
Melanoma is a highly destructive and lethal form of skin cancer. With the contemplation of the concept of plant-based chemotherapeutic and nanomedicine approach, this study aims to develop a liposome of standardized Piper longum L. fruit ethanolic extract (PLFEE), with the evaluation of its pharmaceutical properties and cytotoxic activities against melanoma cell line. To our knowledge, no authentic reports are available for the standardized PLFEE-loaded liposome for melanoma therapy.
Methods:
The chemical marker-based standardization of PLFEE was performed by validated high-performance liquid chromatography (HPLC) to maintain the dosage uniformity and uniform therapeutic outcome. Thin-film hydration was utilized for the formulation of liposomes. The developed liposome was characterized for its pharmaceutical properties and therapeutic activity against melanoma cell lines.
Results:
The prepared liposome was found to be homogeneous, amorphous, transparent, light yellowish, demonstrated nanovesicular/hydrodynamic size (Zavg=104.858±0.262 nm), low polydispersity index (PDI=0.271±0.015), high zeta potential (ζ=-21.8±0.07 mV), spherical morphology, excellent % entrapment efficiency (EE=75.920±3.096%), refractive index (RI=1.335±0.0001), sustained drug release, excellent drug-excipient compatibility, and stability. The liposome showed selective cytotoxicity to B16F10 melanoma cells without affecting healthy HEK293 kidney cells. The PLFEE-loaded liposome showed significantly enhanced cytotoxicity (IC50=59.71±2.364 µg/mL) compared to PLFEE (88.48±3.243 μg/mL).
Conclusion:
Cancer cells use various pathways for their uncontrolled, abnormal proliferation, angiogenesis, invasion, and metastasis. It is, therefore, logical to utilize a multicomponent-based standardized herbal extract that may act through multiple molecular pathways. The liposomal formulation containing multiconstituent-based standardized PLFEE may be a potential alternative chemotherapeutic for melanoma therapy. However, comprehensive in-vivo works are essential to reveal its therapeutic potential.
Keywords: Nanomedicine, Liposomes, Neoplasm, Melanoma, Standardization, Piper
Copyright and License Information
© 2025 The Author(s).
This is an open access article and applies the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
A N Sahu is thankful to the Council of Science and Technology, Uttar Pradesh (CST, UP) for providing funding (Sanction order no: CST/D-1163) for developing carbon nanodots for oral cancer. P C Acharya is thankful to the Department of Biotechnology-Scientific Infrastructure Access for Harnessing Academia University Research Joint Collaboration (DBT-SAHAJ) for providing funding (Sanction order no: BT/INF/22/SP44789/2021) and establishment of Drug Metabolomics Laboratory at Tripura University.
Introduction
Melanoma is a highly aggressive and deadly form of skin cancer.1,2 As per the National Cancer Institute, USA database, its occurrence and mortality rate have been increasing progressively, with 100 640 newly identified cases and 8290 deaths in 2024.3 The majority of the presently used chemotherapeutics have systemic toxicities, adverse effects, narrow therapeutic window, immunity suppression, damage of tissue, and multi-drug resistance.4-7 Finding novel, safe, and effective therapeutics for melanoma is essential for its treatment. From prehistoric times, plant-based therapeutics or phytopharmaceuticals have been used for various therapeutic purposes owing to their diverse chemical structures, safety, and wide therapeutic potential. Several standardized plant extracts, fractions, and bioactives have been well-studied in the preclinical stage for the treatment of melanoma through the regulation of oxidative status, modulation of immunity, correction of disordered replication and induction of apoptosis, prevention of invasion, angiogenesis, and metastasis.8,9 Plant extracts and fractions of Ephedra sinica, Azadirachta indica, Eruca sativa, Tinospora cordifolia, Cordyceps militaris, Viscum album, Calendula officinalis, Andrographis paniculata, Piper longum, Glycyrrhiza glabra, Lawsonia inermis, Solanum lycopersicum, Bauhinia veriegata, Mangifera indica, Withania somnifera, Ganoderma lucidum, Cinnamomum cassia, Inonotus obliquus, Momordica charantia, Allium sativum, Solanum nigrum, Ocimum sanctum, Rhizophora apiculate, Lithospermum erythrorhizon, Zingiber officinale, Decalepis hamiltonii, and Panax ginseng are well reported for their anticancer activity against melanoma through evaluation of various in-vitro and in-vivo tumor regression parameters. Phytoconstituents, such as nimbolide, andrographolide, piperine, piperlongumine, piperlonguminine, isoliquiritigenin, mangiferin, withaferin A, withanolide D, cinnamic aldehyde, alpha momorcharin, ajoene, shikonin, [6]-gingerol, 6-shogaol, ginsenoside Rk, ginsenoside F1, ginsenoside Rg3, ginsenoside Rh2, panaxydol, and resveratrol has been well reported for their in-vitro and in-vivo anticancer activity against melanoma. A detailed list of the standardized extract, fractions, and isolated phytomolecules with their available reports for melanoma therapy has been provided in Supplementary file 1. Compared to isolated constituents, multicomponent-based standardized plant extract may act synergistically for the effective treatment of melanoma in a multi-targeting way.
The Piper longum L. (Family - Piperaceae) extract and its bioactives, mainly the alkaloids, have been well studied to exert anticancer activity against melanoma in preclinical studies.2 Piperine is reported to inhibit various transcription factors and proinflammatory markers in melanoma cells.10 It also causes cell cycle arrest at the G1 phase and induction of apoptosis in melanoma cells.11 Piperine inhibits lung metastasis of melanoma in the C57BL/6 murine model.12 Another alkaloid, piperlongumine is reported to have cytotoxic activity against murine melanoma cells (B16F10) and human melanoma cells (A375, A875). It triggers apoptosis through mitochondrial disruption mediated by reactive oxygen species.13 Likewise, piperlonguminine, an alkaloid, inhibits melanogenesis, offering a rational strategy for treating metastatic melanoma.14 The P. longum L. fruit ethanolic extract (PLFEE) is reported to have antiangiogenic activity.15
Liposomes are the most explored biocompatible nanovesicles, having 20-1000 nm size. They are the most commonly used nanovesicles for delivery of synthetic and natural bioactives due to their capacity to load both hydrophobic & hydrophilic drugs, high biocompatibility, biodegradability, low immunogenicity, targeted drug delivery, protection of drugs from degradation, and clearance. Therefore, considering the concept of a novel plant-based chemotherapeutic and nanomedicine approach, this study aims to develop and characterize a liposomal formulation of standardized P. longum L. extract and evaluate its cytotoxicity for melanoma therapy.
In this research, we describe the formulation of nano vesicular liposomes containing a standardized PLFEE using thin-film hydration, intending to treat melanoma. The rationale behind choosing this nanovesicular formulation was to achieve the optimal therapeutic benefits of PLFEE for melanoma therapy. To date, no such authentic reports have been presented for the treatment of melanoma by the liposome loaded with standardized PLFEE. This will provide opportunities to carry out advanced research on medicinal plants and will develop a platform for the international commercialization of medicinal-plant-based novel therapeutic products for the treatment of melanoma. The chemical marker-based standardization of PLFEE was done utilizing high-performance liquid chromatography (HPLC) to maintain dosage uniformity and uniform therapeutic outcomes. The nanovesicular liposome was characterized for its organoleptic properties, hydrodynamic Zavg, ζ, PDI, pH, loading capacity (% LC), % entrapment efficiency (EE), in- vitro drug release, sedimentation volume, and stability. The morphology of the liposomes was investigated by high-resolution transmission electron microscopy (HRTEM), high-resolution scanning electron microscopy (HRSEM), and scanning probe microscopy (SPM). Further, the liposome was evaluated for crystallinity by selected area electron diffraction (SAED), drug-excipient compatibility by Attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR), and HPTLC. Then the developed liposome was evaluated for cytotoxicity against the melanoma cell line.
Methodology
Materials
Piperine with a purity ≥ 97%, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), fetal bovine serum (FBS), dimethyl sulfoxide (DMSO), and Dulbecco’s modified eagle’s medium (DMEM/F-12) were acquired from Sigma-Aldrich (St. Louis, MO, USA). Piperlonguminine with a purity ≥ 98% was obtained from Cayman Chemical Company (Michigan, USA). Egg lecithin (60% L- ± -phosphatidylcholine) and cholesterol (extrapure AR, 99%) were sourced from Sisco Research Laboratory (Mumbai, India). Absolute ethanol, HPLC grade water and methanol were purchased from Merck Chemicals (Darmstadt, Germany). The dialysis bag (12 000-14 000 Daltons) was obtained from Himedia Laboratories Pvt. Ltd., Mumbai, India. Ultrapure water was employed in the experiments according to the specified requirements.
Plant material collection and authentication
Piper longum L. fruits (Piperaceae family), were procured from a local herbal market and verified by Prof. N.K Dubey (Department of Botany at the Institute of Science, Banaras Hindu University, Varanasi, India), and the voucher specimen (Pipera.2021/6) was submitted to the departmental herbarium. Additionally, the authentication of the obtained fruits involved DNA-based molecular marker identification, as outlined in our previous research. The analysis of nucleotide sequence homology with the ribulose-bisphosphate carboxylase (rbcL) gene was carried out using the Basic Local Alignment Search Tool (BLAST) study offered by the National Institutes of Health (NIH), USA.1,2
Extraction and chemical marker-based standardization by HPLC
Our in-house standardized extract was used for this research work.1,2 The coarsely powdered P. longum L. fruits were extracted using microwave irradiation (450 W, 5 min, microwave oven, HOTBLASTTM, Samsung, South Korea); subsequently, a triple maceration process was implemented using absolute ethanol (powder: solvent-1:4 w/v). The ethanolic extract was filtered, concentrated by a rotary vacuum evaporator (IKA®, RV 10, Germany) at 40 °C, and then kept at 4-8 °C. The PLFEE underwent standardization for piperine and piperlonguminine through a validated HPLC method. Briefly, a measured quantity of the standardized PLFEE was combined with ethanol, subjected to vortexing and bath sonication (10 minutes), and then syringe filtered (0.2 μm). Then, the obtained filtrate was suitably diluted with HPLC-grade methanol. Subsequently, 20 μL of the prepared sample was introduced into the HPLC for quantification of piperine and piperlonguminine. The quantification was performed using the validated HPLC method, with detection wavelengths set at 342 nm for piperine and 340 nm for piperlonguminine.1,2
HPLC method development
HPLC was carried out utilizing an Agilent 1260 Infinity II system equipped with an autosampler, quaternary pump, reverse phase C18 column (PerkinElmer, 250 × 4.6 mm, 5 μm), and photodiode array detector. The elution was carried out at a flow rate of 1.00 mL/min using HPLC-grade methanol and water at a ratio of 80:20 v/v for 10 minutes run time. Then the method was validated as per International Conference on Harmonization Tripartite (ICH) guidelines.16 The validation encompassed key parameters such as linearity, range, accuracy (% recovery), precision (including intermediate precision and repeatability), limit of detection (LOD), limit of quantification (LOQ), system suitability, and robustness. A comprehensive description of the validation procedure is described in our previous publication and other referenced sources.1,2
Preparation of liposomes
The liposomes loaded with PLFEE were prepared by thin-film hydration.17 The required amount of PLFEE, egg lecithin (EL), and cholesterol (CHOL) (as given in Table 1) were dissolved properly in absolute ethanol separately and then mixed in a 100 mL round-bottom flask for 10 min by shaker orbital shaking lubricator (REMI, RS 12 plus). Then, using a rotary vacuum evaporator (IKA®, RV 10, Germany) the ethanol was evaporated at 40 °C, 60 RPM, till the formation of a thin film inside the flask. The trace amount of solvent was removed by vacuum for 5 hours. Then, the formed film was hydrated with ultrapure water to prepare liposome vesicles. Vesicle size reduction was achieved by homogenizer (Ultra-Turrax T25 Homogenizer, IKA, Inc, Willington, NC, USA) at 3400 RPM for 5 minutes. The unentrapped extract was removed by ultracentrifugation at 100 000 g using Beckman Optima XPN-100 for 1 hour at 4 °C, discarding the supernatant. Then, the pellet was reconstituted by ultrapure water to form liposomes. The liposomal formulations were sterilized by filtration using a Pall-Gelman Supor Acrodisc® syringe filter (0.22 µm), kept in a screw-capped glass vial, and stored protected from light at 4 °C. Piperine was used as a representative marker throughout the study.
Table 1.
Composition of nano liposomal formulations containing ethanolic extract of Piper longum L.
|
Sl No.
|
Formulation Codes
|
PLFEE (mg)
|
EL (mg)
|
CHOL (mg)
|
| 1 |
LF1 |
50 |
50 |
5 |
| 2 |
LF2 |
50 |
100 |
10 |
| 3 |
LF3 |
50 |
150 |
15 |
| 4 |
LF4 |
50 |
200 |
20 |
PLFEE: Piper longum L. fruits ethanolic extract; EL: Egg lecithin; CHOL: Cholesterol.
Organoleptic properties
The prepared liposomal formulations were evaluated for colour, clarity, homogeneity, transparency, etc.
Zavg, PDI, and ζ
The hydrodynamic Zavg, their PDI, and ζ of the liposomes were assessed using a Nano ZS 90 Zetasizer (Malvern, UK)18 using disposable cells. Each liposomal formulation underwent a 10-fold dilution with ultrapure water and filtration using a 0.22 µm syringe filter (Pall-Gelman Supor Acrodisc®) to prevent multiple scattering effects.
pH
The pH of the liposome was measured using a PC 700 digital pH meter (Eutech, Singapore). Each formulation was diluted with a 10-fold Millipore water, and pH measurements were conducted at room temperature.
Refractive index (RI)
The RI of all the liposomes was estimated to verify the isotropy of the formulations by estimating and comparing the RI of samples collected from three different regions of each liposomal formulation using an Abbe-type refractometer (Labman, India) at 25 °C. A few drops of undiluted liposome were evenly applied to the lower illuminating prism, and the analysis was conducted in triplicate.
EE and LC
The ultracentrifugation method was used to estimate the EE and LC.19 Accurately, 6 mL of each formulation was ultracentrifuged using Beckman Optima XPN-100 ultracentrifuge (Palo Alto, CA, USA) at 100 000 g at 4 °C for 1 hour. The total quantity of piperine in the liposomes before centrifugation and free piperine after centrifugation present in the supernatant was estimated by validated HPLC method after suitable dilution with HPLC grade methanol. Then, the EE was calculated using Eq.1.
Where EE is the entrapment efficiency, TP is the total amount of piperine in the taken volume of sample, and FP is the amount of free piperine after centrifugation present in the supernatant.
The LC of liposomes was calculated as per Eq.2.
Where LC is the loading capacity, TP is the total amount of piperine in the taken volume of sample, FP is the amount of unbound piperine in the supernatant post-centrifugation, and TE is the total quantity of excipient incorporated in the liposomal formulation.
Sedimentation volume
Five milliliters of each prepared liposomal formulation was transferred to a 10 mL volume measuring cylinder and allowed to settle for 10 days at 25 °C. The cylinders were observed at the end of each day, and the sedimentation volume was calculated using Eq.3.
(3)
Where Vu is the ultimate volume of sedimentation, and V0 is the total volume of liposomal dispersion.
HRSEM
The 3D morphology of the optimized liposome was assessed using a Nova Nano SEM 450 instrument (FEI, USA) equipped with an Everhart-Thornley detector (ETD) at 15 kV. In this study, 60 µL of the formulation was applied to 1 cm2 of aluminium foil and allowed to dry, resulting in a dry film. This dried film was affixed to a metal stub and coated with gold using a DSR 1 sputter coater (Nanostructured Coating Co., Iran) for 120 seconds. The vesicle size was analyzed using ImageJ software (NIH, Maryland), and the average size was estimated from the analysis of 25 vesicles.
SPM
SPM analysis was conducted utilizing the NTEGRA Prima microscopy system (NT-MDT Service and Logistics Ltd.). The optimized liposome was placed on a 1 cm2 clean glass, allowed to air-dry, and then subjected to the instrument for both 2D and 3D morphological analysis.
HRTEM and SAED analysis
The 2D morphology assessment of liposomal vesicles and the analysis of the SAED pattern were performed using a FEI TECNAI G2 20 TWIN instrument (USA) at 200 kV. The prepared liposome was diluted tenfold with ultra-pure water, filtered using a 0.22 µm syringe filter dropped onto a carbon-coated copper grid (400 Mesh, Agar Scientific). The excess sample was removed using soft tissue paper, and the grid was left to air-dry at room temperature. The dried TEM grid was then subjected to HRTEM for morphology analysis.
Drug-excipient compatibility study
The ATR-FTIR and HPTLC were used and studied for evaluating drug-excipient compatibility.
ATR-FTIR
The ATR-FTIR study was carried out using Bruker Alpha II ATR-FTIR spectrophotometer (Germany). Since the liposome is liquid, to avoid the broad peak of water, it was freeze-dried for 12 hours using an LFD-BT-101 freeze dryer (Labocon Scientific Limited, UK). A small quantity of PLFEE, EL, CHOL, physical mixture, and freeze-dried liposomal formulation (LF3) was uniformly spread on the ATR crystal on which the probe was fixed tightly. Spectra was recorded with a 4 cm-1 spectral resolution and 40 scans within the wave number of 4000-600 cm-1.
HPTLC
The fingerprinting of ethanolic extract and liposomal formulation was conducted using an HPTLC instrument (CAMAG) equipped with WinCATS software with a 20 × 10 cm pre-coated silica gel 60 F254 plate (Merck, Germany). Briefly, the samples were dissolved in ethanol, centrifuged at 10 000 rpm for 5 minutes, and the supernatant was collected. Accurately 5 μL of each sample was applied in triplicate with 8 mm bandwidth using a Linomat V applicator fitted with a 100 μL syringe. Post application, the plate was developed in a pre-saturated twin trough glass chamber up to 8 cm using 20 mL of toluene: ethyl acetate (7:3 v/v) as mobile phase.20 After drying, the plate was scanned by an HPTLC scanner IV at a speed of 20 mm/s at 254, 366, and 412 nm. The retardation factor (Rf) of the sample and pure piperine bands were determined, and drug-excipient compatibility was assessed by comparing Rf values.21 The developed fingerprint was visualized under TLC Visualizer 2.
In-vitro drug release and release kinetics study
The release study was conducted using piperine as a representative marker. A dialysis bag (Dialysis Membrane-70, MWCO 12-14 kDa) was immersed overnight in deionized water to eliminate preservatives. The release pattern of PLFEE-loaded liposome was compared with the PLFEE solution (PBS with 0.5% Tween 80), and PLFEE suspension (PBS only) as control samples. Two milliliters of liposomal formulation, non-formulated PLFEE solution, and PLFEE suspension were added into a dialysis bag and dipped in a 50 mL release medium (pH 7.4 PBS with 0.5% Tween 80) in a beaker. The release medium was maintained at 37°C ± 0.5 °C with stirring at 150 RPM using a temperature-controlled magnetic stirrer with a hot plate (IKA C-MAG HS 7). At specific intervals (0, 0.083, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5 6, 8, 10, 12, 14, 16, 18, 20, 22, and 24 hours), 1 mL of the release sample was withdrawn and replenished with fresh release medium to achieve sink conditions. The HPLC method was employed to quantify the released piperine, and the results were graphically expressed as the cumulative percentage of drug released over time. Mathematical kinetic models, including zero order, first order, Korsmeyer-Peppas, Higuchi, and Hixson-Crowel cube root law, were fitted to the in-vitro drug release data. Linear regression analysis was performed, and the correlation coefficient (R2) was calculated in each case.
Cytotoxicity assay
The cytotoxicity of the nano liposomal formulation and the free standardized extract was carried out against melanoma cells (B16F10) using the MTT assay as per our reported protocol.1,2 Noncancerous human embryonic kidney cells (HEK 293) were used for comparison. Briefly, the cell lines were obtained from the National Centre for Cell Science (NCCS), Pune, India, and seeded at a concentration of 1 × 106 cells/well in 96 well plates containing DMEM/F-12 medium with 10% FBS, 50 unit/mL penicillin, and streptomycin. The well was incubated in a 5% CO2 atmosphere for 24 hours at 37 °C. Then, the medium was discarded, washed thrice with phosphate-buffered saline (pH 7.4), and incubated with serum-free fresh media containing different concentrations (0-160 µg/mL) of test substances (PLFEE solution in 0.2% DMSO and PLFEE-loaded liposome) and negative control (placebo liposome and 0.2% DMSO) for 24 hours. Then the cells are washed with PBS and treated with 200 μL of MTT solution (5 mg/mL), and further incubated for 4 hours at 37 °C. The formazan crystals developed from MTT reduction were dissolved by the addition of 150 µL of DMSO. The absorbance was recorded using a SpectraMax M5 microplate reader (Molecular Devices, USA) at 570 nm. Then, the percentage cell viability was estimated using Eq.4.
(4)
Where, Abs(t) and Abs(c) are the absorbances of formazan with the treatments and the control, respectively. The inhibition concentration (IC50) was assessed from nonlinear regression using GraphPad Prism (San Diego, California). The % cell viability at each concentration of standardized PLFEE and liposome were compared at P < 0.05 via paired student t-test.
Stability study
Various stability-indicating parameters, such as colloidal dispersion stability (Zavg, PDI, and ζ), physical properties (colour, pH, RI, sedimentation, precipitation, and phase separation), chemical integrity (FTIR), and pharmaceutical properties (% EE and % LC) were accessed intermittently and at the end of the study.
Colloidal dispersion stability
The liposomal formulations were kept at 4 °C and 25 °C for 90 days, with periodic evaluations of Zavg, PDI, and ζ every 15 days.
Physical stability
Physical stability assessments were conducted at different time intervals by storing the samples at 4 °C and 25 °C for a period of 90 days. The inspection of physical characteristics, including colour, pH, RI, sedimentation, phase separation, and precipitation, was performed at 15-day intervals.
Chemical stability
The chemical stability was evaluated in terms of chemical integrity by ATR-FTIR and pharmaceutical possessions (% EE and % LC) by validated HPLC. The liposomal samples were kept under 4 °C & 25 °C for 90 days, and the stability was accessed at an interval of 15 days.
Thermal stability
To evaluate thermal stability, the sample underwent heating-cooling and freeze-thaw cycles. The stability assessment included three cycles of heating-cooling, each lasting 24 hours at 40 °C and 4 °C, respectively, as well as three cycles of freeze-thaw, each lasting 24 hours at -20 °C and 25 °C.
Lyophilization stability
The lyophilization stability was studied to observe possible changes in colloidal and pharmaceutical properties of the liposome using trehalose as a cryoprotectant. The freshly prepared liposome was subjected to pre-freezing at -80 °C for 12 h, followed by lyophilization in a lyophilizer (LFD-BT-101, Labocon Scientific Limited, UK). The Zavg, PDI, ζ, % EE, and % LC were accessed after reconstitution with ultrapure water.
Statistical analysis
All the studies were performed in triplicate, and the outcomes are presented as mean ± standard deviation. The Statistical analysis was carried out using a one-way analysis of variance (ANOVA) followed by Tukey’s test and Student’s t-test at a significance level of P < 0.05, utilizing GraphPad Prism 5.
Results
HPLC methodology and validation
Both the chemical markers (piperine and piperlonguminine) showed the linearity within 2 to 30 µg/mL with a linear equation y = 276947.424 x - 200378.812 (n = 3, r2 = 0.997) for piperine and y = 349329.568 x - 272061.184 (n = 3, r2 = 0.996) for piperlonguminine. The complete validation outcomes are reported in our previous reports.1,2 Briefly, the high correlation coefficient values (r2 > 0.99) in both cases indicate a strong relationship among the variables. The results of the accuracy are represented in terms of % recovery and percentage relative standard deviation (% RSD). The high recovery values and low percent RSD ( < 2%) reflected the achievement of excellent accuracy. Precision indicates the degree of closeness of agreement among a series of readings under specified analytical conditions. The repeatability and intermediate precision (intra-day and inter-day) studies were performed to examine the precision of the HPLC method. The % RSD values are found to be less than 2%, which fulfilled the acceptance criteria and showed a high degree of precision of the developed HPLC method. The LOD and LOQ values for the determination of piperine were found to be 0.259 and 0.786 μg/mL, respectively. Similarly, the LOD and LOQ values for piperlonguminine were found to be 0.27 and 0.819 μg/mL, respectively. Such lower values of LOD and LOQ of the developed HPLC method indicated the sensitivity of the optimized method. Robustness is a measure of the ability of the HPLC method to remain unaffected by slight but deliberate changes in the method variables that offer its reliability during normal usage. The robustness of the developed HPLC method investigated by deliberate changes in different chromatographic conditions showed the lower % RSD values ( < 2%) of the area and retention time (RT), which confirm the robustness of the developed HPLC method. The system suitability checks the specificity and validity of the developed analytical method. The % RSD of various validation parameters, RT, peak area, the number of theoretical plates, peak purity, tailing factor, and capacity factor were estimated for system suitability analysis. The % RSD of all examined parameters is found to be within the permissible limits ( < 2%). The number of theoretical plates (N > 2000) and tailing factor ( < 2) were within the acceptable limit. The peak purity of piperine and piperlonguminine was found to be 1, representing a high degree of purity. The capacity factor (K1) represents the degree of interaction of the analyte with the stationary phase in the column. A capacity factor of zero indicates no interaction, whereas a capacity factor > 1 indicates a certain degree of interaction with the stationary phase. In both cases, the K1 values are found to be within the optimal range (1-5), thus requiring less run time (10 minutes) for analysis. The system suitability results disclose the suitability of the developed HPLC system for quantitative analysis of piperine and piperlonguminine. The overall validation outcomes reflected the suitability of the developed HPLC method for accurate quantification of the potential markers in the extract.1,2
Extraction and chemical marker-based standardization by HPLC
The taxonomical and DNA-based molecular authentication was carried out to confirm the authenticity of the P. longum L. fruits (Family: Piperaceae).2 The extraction yield was 16.532%. The amounts of piperine and piperlonguminine were 377.0687 ± 1.453 mg and 6.72 ± 0.108 mg per gram of dried PLFEE.
Formulation and development of liposome
The graphical representation of the formulation method for liposomes is depicted in Figure 1.

Figure 1.
The formulation scheme of PLFEE-loaded liposome
.
The formulation scheme of PLFEE-loaded liposome
Organoleptic properties
The liposomes were found to be clear, homogeneous, transparent, and light yellowish without any sedimentation of particulate materials at the bottom of the vials (Figure 1).
Zavg, PDI, and ζ
The outcomes of various characterization parameters of the prepared PLFEE-loaded liposomes are shown in Table 2. The Zavg & PDI were represented in Figure 2a, and ζ was shown in Figure 2b. The average particle size of prepared liposomes ranged from 94.193 ± 0.25 to 115.395 ± 0.086 nm, representing the nanovesicular size (Table 2, Figure 2).
Table 2.
Characterizations of PLFEE-loaded liposomes
|
Formulation codes
|
Z
avg
(nm)
|
PDI
|
ζ (mV)
|
pH
|
RI
|
EE (%)
|
LC (%)
|
| LF1 |
94.193 ± 0.25 |
0.253 ± 0.002 |
-20.1 ± 0.31 |
4.586 ± 0.275 |
1.334 ± 0.0001 |
56.357 ± 2.124 |
9.043 ± 0.121 |
| LF2 |
103.563 ± 0.319 |
0.254 ± 0.002 |
-30.0 ± 0.29 |
4.43 ± 0.1 |
1.335 ± 0.0002 |
64.124 ± 1.154 |
6.397 ± 0.213 |
| LF3 |
104.858 ± 0.262 |
0.271 ± 0.015 |
-21.8 ± 0.071 |
4.266 ± 0.045 |
1.335 ± 0.0001 |
75.920 ± 3.096 |
4.943 ± 0.341 |
| LF4 |
115.395 ± 0.086 |
0.273 ± 0.005 |
-17.8 ± 0.831 |
4.3 ± 0.05291 |
1.335 ± 0.0003 |
85.983 ± 2.027 |
4.142 ± 0.273 |
Values represented as mean ± SD (n = 3).
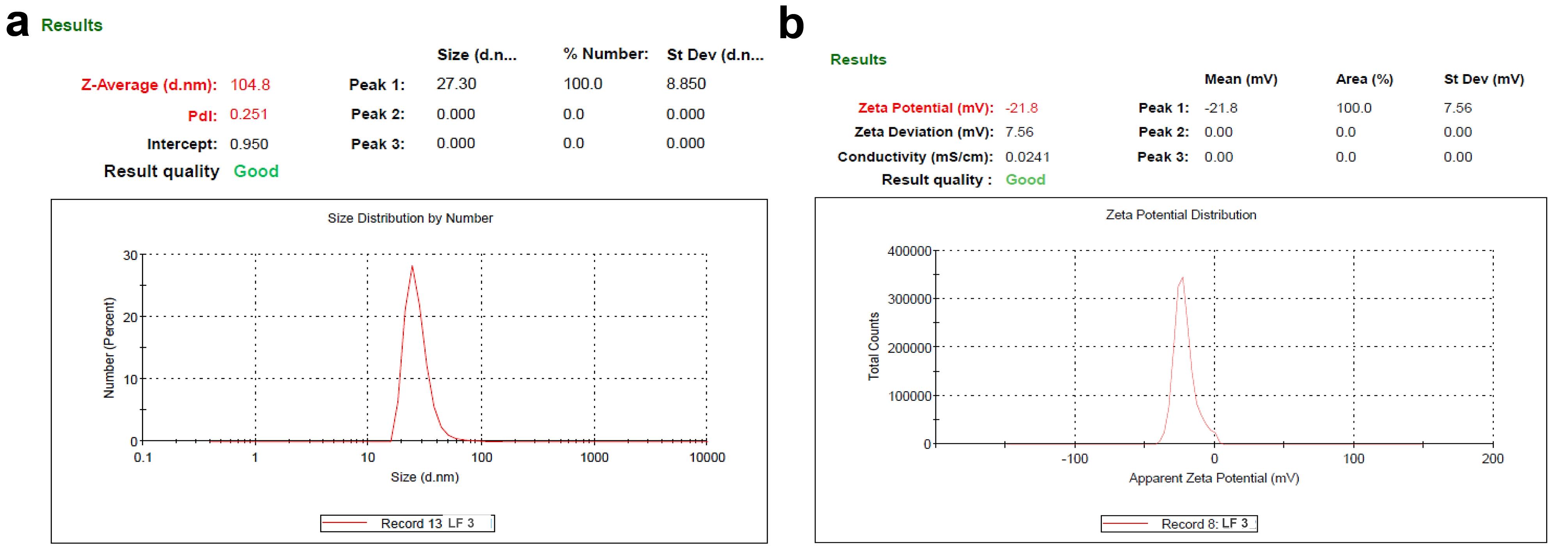
Figure 2.
Zavg and ζ of PLFEE-loaded (LF3) (a) average Zavg and (b) ζ
.
Zavg and ζ of PLFEE-loaded (LF3) (a) average Zavg and (b) ζ
pH and RI
The results of pH and RI are shown in Table 2. All the liposomal formulations show an acidic pH range between 4.266 ± 0.045 to 4.586 ± 0.275. RI of the liposomes are found to be between 1.334 ± 0.0001 to 1.335 ± 0.0003.
EE and LC
The % EE and % LC of various batches of liposomal formulation are shown in Table 2. The value of % EE was found to be within 56.357 ± 2.124 to 85.983 ± 2.027%, and % LC was found to be within 4.142 ± 0.273 to 9.043 ± 0.121%.
Sedimentation volume
The sedimentation volumes of prepared liposomes evaluated each day and at the end of 10 days are found to be zero.
HRSEM
The PLFEE-loaded LF3, which has intermediate entrapment efficiency, vesicle size, PDI, and ζ, was considered for surface morphology analysis. The photomicrograph obtained from HRSEM analysis is shown in Figure 3a. The size distribution graph of 25 vesicles analyzed by ImageJ is shown in Figure 3b. The liposomal vesicles are found to be spherical shaped with nanometric size within 85 to 115 nm and an average size of 100.665 nm.

Figure 3.
HRSEM and SPM photomicrographs of PLFEE-loaded LF3 (a) HRSEM photomicrograph (b) Size distribution of 25 vesicles analyzed by ImageJ software, (c) 2D SPM photomicrograph, and (d) 3D SPM photomicrograph
.
HRSEM and SPM photomicrographs of PLFEE-loaded LF3 (a) HRSEM photomicrograph (b) Size distribution of 25 vesicles analyzed by ImageJ software, (c) 2D SPM photomicrograph, and (d) 3D SPM photomicrograph
SPM
The 2D photomicrograph (Figure 3c) of LF3 vesicles is found to have spherical and smooth surfaces with vesicle sizes closer to 100 nm. The 3D SPM photomicrograph (Figure 3d) demonstrated the height of the liposomal vesicles to be closer to 100 nm.
HRTEM and SAED
The HRTEM photomicrograph of PLFEE-loaded LF3 presented in Figure 4a confirms the regular, uniform, spherical shape of the liposomal vesicles, with most of the particles in the size ranges between 84.48 to 103.34 nm. The SAED patterns in Figure 4b show a diffused ring.
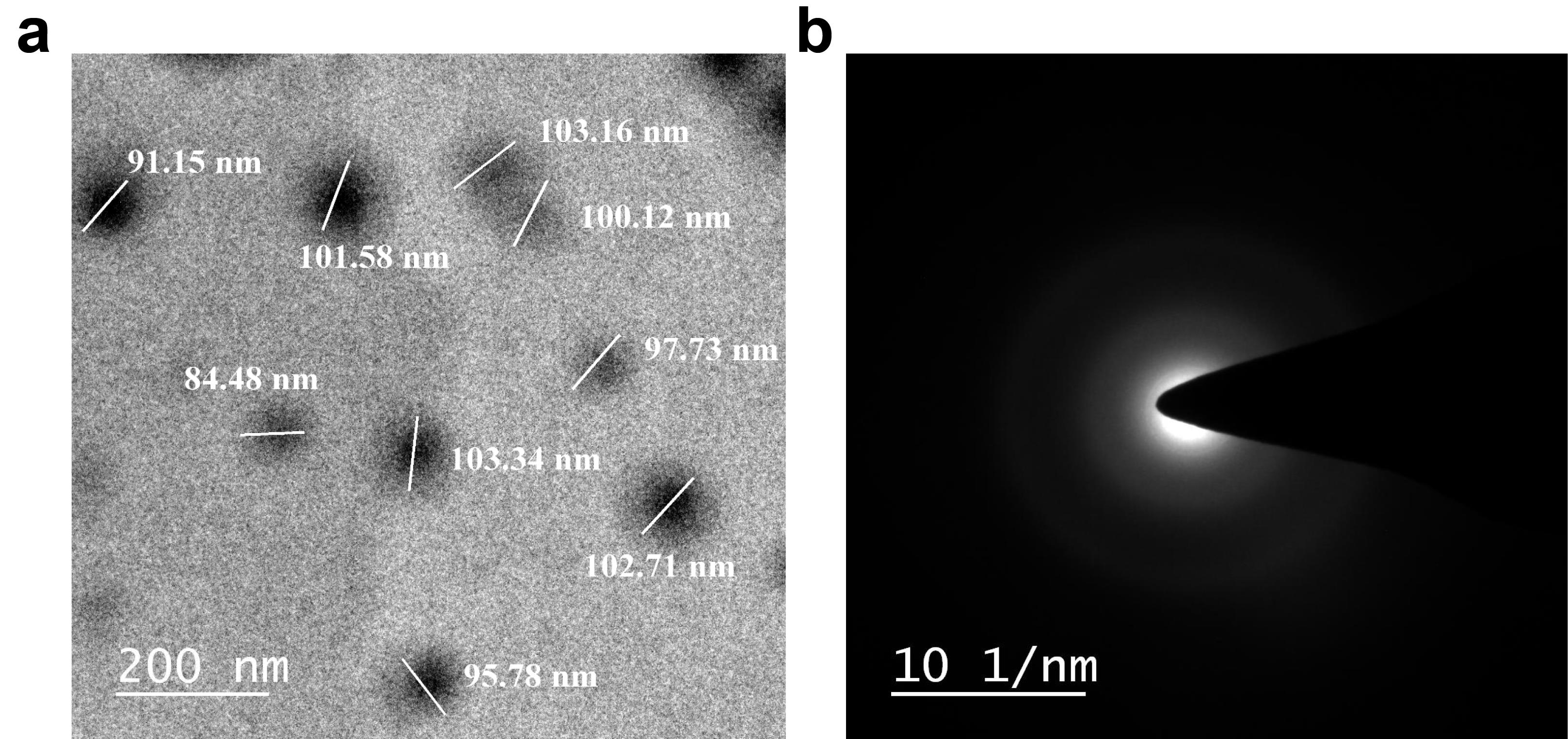
Figure 4.
HRTEM Photomicrograph and SAED pattern of PLFEE-loaded liposome (LF3) (a) HRTEM photomicrograph of LF3 at 200 nm scale, and (b) SAED pattern of PLFEE-loaded LF3. HRTEM: High-resolution transmission electron microscopy; SAED: Selected area electron diffraction
.
HRTEM Photomicrograph and SAED pattern of PLFEE-loaded liposome (LF3) (a) HRTEM photomicrograph of LF3 at 200 nm scale, and (b) SAED pattern of PLFEE-loaded LF3. HRTEM: High-resolution transmission electron microscopy; SAED: Selected area electron diffraction
Drug-excipient compatibility study
ATR-FTIR
The results of the drug-excipient compatibility investigation, as assessed through ATR-FTIR and HPTLC, are presented in Figure 5. Standardized PLFEE exhibited distinctive vibrational peaks (Figure 5a) due to C-H stretching at 2925.48 cm-1 and 2851.53 cm-1, -C = O-N = stretching at 1633.57 cm-1, aliphatic diene stretching at 1613.43 cm-1, C = C aromatic stretching at 1442.65 cm-1, = C-O-C stretching at 1245.75 cm-1, = C-O-C symmetrical stretching at 1033.43 cm-1, C–H bending at 995.75 cm-1, and C-O stretching at 927.46 cm-1. These results align with previous FTIR findings on PLFEE.1,2
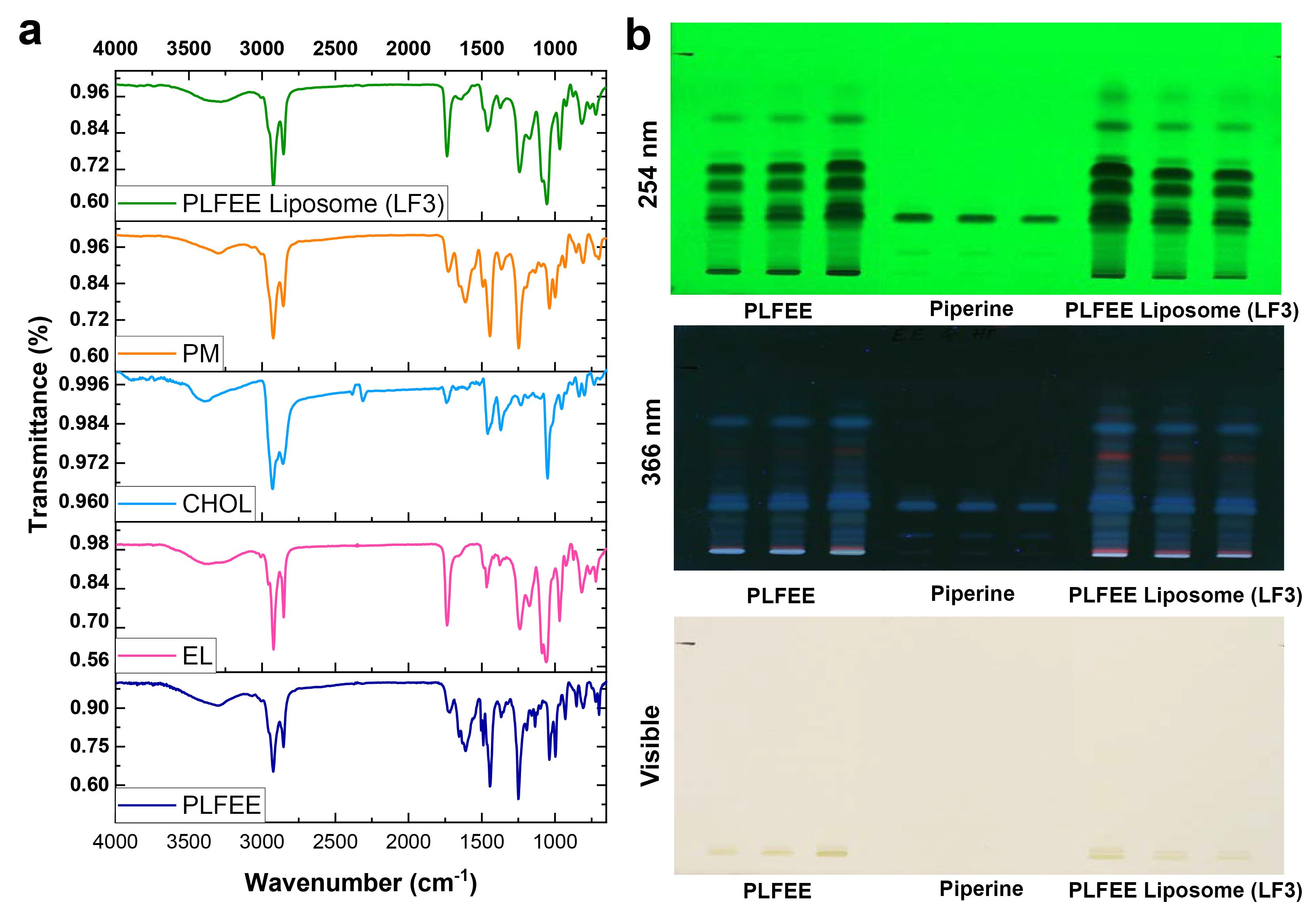
Figure 5.
Drug-excipient compatibility study by (a) ATR-FTIR spectra of standardized extract, formulation components, physical mixture of formulation components, and PLFEE-loaded LF3, and (b) HPTLC fingerprint of PLFEE, Piperine, and PLFEE-loaded LF3. PM: Physical mixture; LF: Liposomal formulation
.
Drug-excipient compatibility study by (a) ATR-FTIR spectra of standardized extract, formulation components, physical mixture of formulation components, and PLFEE-loaded LF3, and (b) HPTLC fingerprint of PLFEE, Piperine, and PLFEE-loaded LF3. PM: Physical mixture; LF: Liposomal formulation
EL showcased a broad peak due to -OH stretching at 3387.142 cm-1, C-H stretching due to methylene group at 2923.506 cm-1 and 2849.697 cm-1, C = O stretching at 1736.092 cm-1, C-H bending at 1464.976 cm-1, P = O stretching at 1240.538 cm-1, and P-O-C stretching at 1064.75 cm-1.22
CHOL exhibited a broad peak due to stretching of the -O-H group at 3383.150 cm-1, asymmetric stretching of C–H bonds at 2925.983 cm-1 and 2852.862 cm-1, C = C stretching or C–H bending at 1464.976 cm-1, C-O stretching of C-O-H group at 1365.783 cm-1, O-H bending at 1221.354 cm-1, C-O stretching at 1042.843 cm-1, and C–C backbone vibration at 947.243 cm-1 and 834.852 cm-1, along with out-of-plane vibrations of C–H group at 793.351 cm-1.23
The ATR-FTIR spectrum of the physical mixture (PM) is a composite of standardized PLFEE, EL, and CHOL. The freeze-dried PLFEE-loaded LF3 predominantly exhibited all the characteristic peaks of standardized PLFEE.
HPTLC
The study of the compatibility of the standardized PLFEE with formulation excipients was conducted using HPTLC fingerprinting. The fingerprints of PLFEE, piperine, and the PLFEE-loaded LF3 are depicted in Figure 5b. Among the various experimented mobile phases, toluene: ethyl acetate (7:3 v/v) yielded optimal results at a retardation factor (Rf) of 0.26 ± 0.0054 for piperine in the pure piperine sample, ethanolic extract, and liposomal formulation (Figure 5b). Additionally, all the chromatographic bands observed in neat PLFEE were retained in the PLFEE-loaded liposomal formulation.
In-vitro release and release kinetics
Figure 6a illustrates the in-vitro release profiles of PLFEE solution, PLFEE suspension, and PLFEE-loaded LF3. In the initial 4 hours, the PLFEE solution exhibited a rapid drug release (85.043 ± 3.124%), while the PLFEE liposome demonstrated sustained release at 4 hours (40.86 ± 2.175%), followed by the PLFEE suspension (15.376 ± 2.124%). After 6 hours, the release of the PLFEE solution reached a plateau, while the PLFEE-loaded liposome continued releasing the drug for up to 24 hours, demonstrating a biphasic release pattern. The liposomes showed Higuchi diffusion kinetics among the various kinetic models (Table 3) due to the highest value of linear regression coefficient (R2 = 0.963).
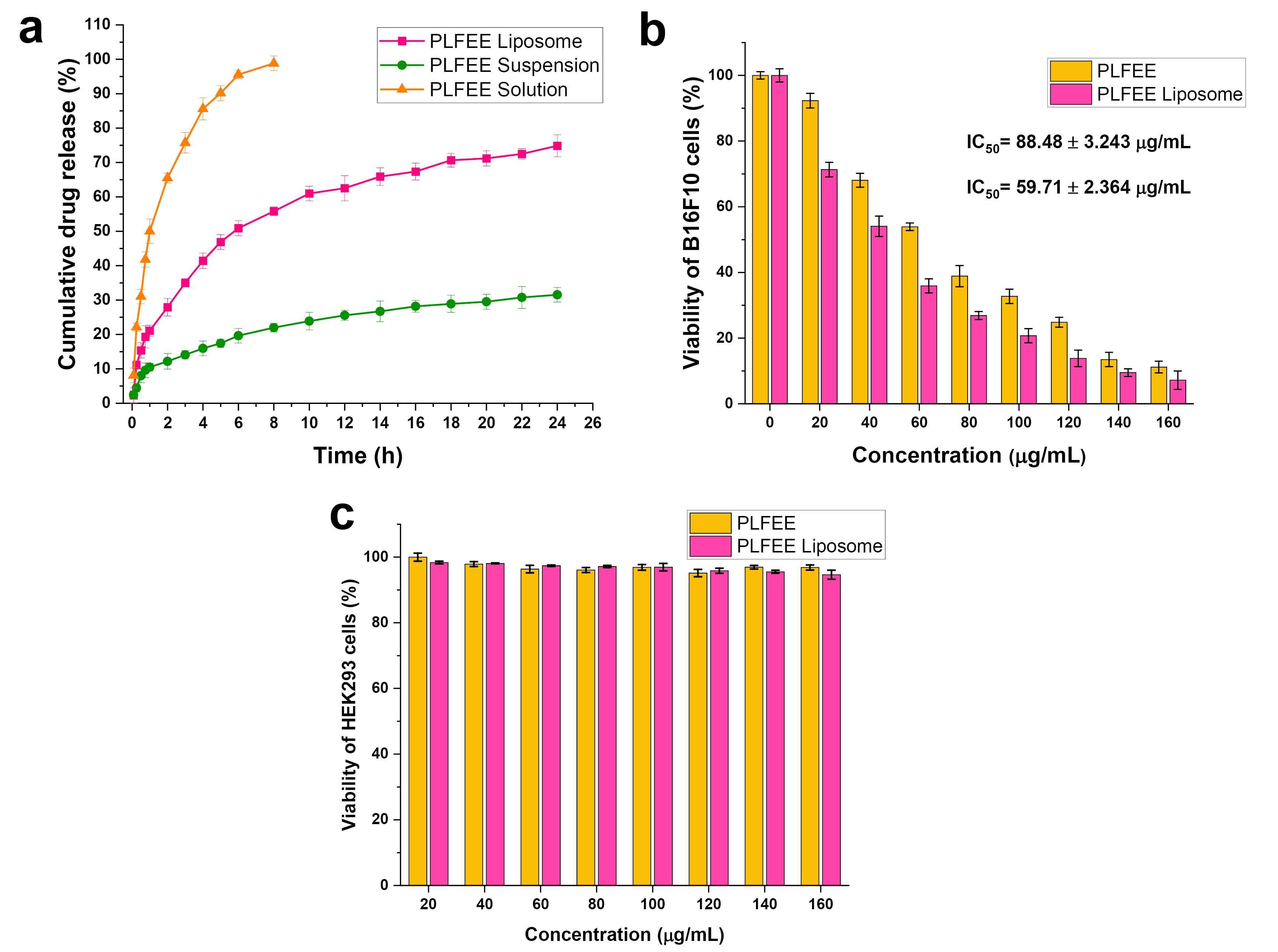
Figure 6.
In-vitro drug release and cytotoxicity studies (a) Comparative in-vitro release profile of PLFEE liposome, suspension, and solution through dialysis bag, (b) Cytotoxicity of PLFEE and PLFEE loaded liposome against B16F10 after 24 h, and (c) Cytotoxicity of PLFEE and PLFEE loaded liposome against HEK293 after 24 h. B16F10: Metastatic melanoma cells; HEK 293: Human embryonic kidney 293 cells
.
In-vitro drug release and cytotoxicity studies (a) Comparative in-vitro release profile of PLFEE liposome, suspension, and solution through dialysis bag, (b) Cytotoxicity of PLFEE and PLFEE loaded liposome against B16F10 after 24 h, and (c) Cytotoxicity of PLFEE and PLFEE loaded liposome against HEK293 after 24 h. B16F10: Metastatic melanoma cells; HEK 293: Human embryonic kidney 293 cells
Table 3.
Kinetics of drug release from PLFEE-loaded liposome
|
Zero-order
|
First-order
|
Higuchi Model
|
Korsmeyer-Peppas model
|
Hixson-Crowell model
|
Mechanism of drug release
|
|
K0
|
R2
|
K1
|
R2
|
KH
|
R2
|
Kkp
|
R2
|
KHC
|
R2
|
n
|
R2
|
| 2.682 |
0.835 |
0.023 |
0.933 |
15.27 |
0.963 |
0.515 |
0.929 |
0.081 |
0.664 |
0.598 |
0.931 |
K0: Zero-order release constant; K1: First-order rate constant; KH: Higuchi constant; kkp: Korsmeyer rate constant; and KHC: Hixson-Crowell rate constant; n: release exponent, and R2: Regression coefficient.
Cytotoxicity assay
The in-vitro cytotoxicity results of standardized PLFEE and PLFEE-loaded LF3 against melanoma (B16F10) and healthy kidney cells (HEK293) after 24 hours of treatment are represented in Figure 6b and Figure 6c, respectively. The calculated IC50 values for neat PLFEE and PLFEE-loaded LF3 against B16F10 are 88.48 ± 3.243 µg/mL (R2 = 0.996) and 59.71 ± 2.364 µg/mL (R2 = 0.996), respectively. Particularly, the standardized extract and extract-loaded liposome demonstrate non-toxicity to normal HEK293 cells (Figure 6c).
Stability of prepared liposomes
The results of the stability study of PLFEE-loaded LF3 are shown in Table 4.
Table 4.
Stability study results of liposome at the end of the stability study
|
Parameters
|
Initial results
|
Under refrigerator
|
At room temperature
|
Freeze-thaw cycle
|
Heating-cooling cycle
|
Lyophilization stability
|
| Zavg (nm) |
104.858 ± 0.262 |
110.324 ± 1.233 |
145.347 ± 3.124 |
107.258 ± 1.114 |
124.357 ± 3.257 |
103.357 ± 1.234 |
| PDI |
0.271 ± 0.015 |
0.289 ± 0.013 |
0.764 ± 0.0234 |
0.274 ± 0.016 |
0.441 ± 0.035 |
0.273 ± 0.005 |
| ζ (mV) |
-21.8 ± 0.071 |
-20.57 ± 0.234 |
-16.43 ± 0.335 |
-21.31 ± 0.123 |
-19.47 ± 0.087 |
-21.14 ± 0.036 |
| Color |
Light yellow |
Light yellow |
Light yellow |
Light yellow |
Light yellow |
Light yellow |
| pH |
4.266 ± 0.045 |
4.324 ± 0.034 |
4.224 ± 0.023 |
4.273 ± 0.033 |
4.235 ± 0.044 |
4.254 ± 0.033 |
| RI |
1.335 ± 0.0001 |
1.335 ± 0.002 |
1.334 ± 0.0003 |
1.335 ± 0.001 |
1.334 ± 0.004 |
1.335 ± 0.0002 |
| Sedimentation |
No |
No |
Slight sedimentation after 15 days |
No |
Slight sedimentation after 3rd cycle |
No |
| Precipitation |
No |
No |
No |
No |
No |
No |
| Phase separation |
No |
No |
Slight separation after 15 days |
No |
Slight separation after 3rd cycle |
No |
| Chemical integrity |
Retained |
Retained |
Retained |
Retained |
Retained |
Retained |
| EE (%) |
75.920 ± 3.096 |
72.354 ± 2.157 |
55.753 ± 3.254 |
75.125 ± 1.258 |
73.753 ± 2.157 |
73.357 ± 2.234 |
| LC (%) |
4.943 ± 0.341 |
4.667 ± 0.951 |
2.753 ± 1.256 |
4.914 ± 0.287 |
3.143 ± 0.348 |
4.568 ± 0.573 |
Colloidal dispersion stability
The Zavg and PDI were found to be maintained for up to 90 days with a slight increase in the values under refrigerator conditions; however, it was drastically increased when stored at room temperature (Table 4). The higher PDI value (0.764 ± 0.0234) at room temperature represented the heterogeneous nature of the vesicles with broad size distribution. The ζ was found to be slightly decreased with the lapse of time under a refrigerator and greatly reduced under room temperature.
Physical stability
The results of physical stability represent no substantial changes in the physical properties (Color, pH, RI, sedimentation, precipitation, and phase separation) under refrigerated conditions for up to 3 months. In contrast, slight sedimentation and phase separation were observed at room temperature.
Chemical stability
The chemical integrity, as assessed through ATR-FTIR spectra, was found to be maintained throughout the stability study. No distinguished changes were observed in the percentages of encapsulation efficiency (% EE) and loading capacity (% LC) under refrigeration conditions. However, a significant decrease was observed under room temperature.
Thermal stability (heating-cooling cycle & freeze-thaw cycle)
The Zavg, PDI, and ζ were found to be maintained at the end of the freeze-thaw cycle, whereas it was found to be significantly changed at the end of the heating-cooling cycle, which might be due to agglomeration of liposomal vesicles. No substantial changes in the physical properties, % EE, and % LC were observed after the freeze-thaw cycle, whereas slight sedimentation, phase separation, and a slight decrease in % EE and % LC were observed after 3rd cycle of the heating-cooling cycle. The chemical integrity was retained throughout the thermal stability study.
Lyophilization stability
Upon reconstitution of the lyophilized liposomes, the values of Zavg, PDI, and ζ were found to closely resemble the initial values of the liposomes. Additionally, after the lyophilization study, no significant variations in the physical and chemical properties were noticed.
Discussion
The preparation of PLFEE-loaded liposomes utilized the thin film hydration method owing to its simplicity, ease of monitoring processing variables such as rotation and temperature, and scalability. Moreover, this method permits the formation of a thin film over a huge surface area and facilitates the effective hydration of nanovesicles, thereby enhancing entrapment efficiency.1 Ethanol was selected for extraction and formulation development due to its safety (Class III solvent) and excellent solubilizing ability of formulation components. The EL acts as a membrane former and constitutes the bilayer structure of the liposome. CHOL improves the rigidity and stability of liposomal vesicles. The incorporation of CHOL in the liposomal bilayer modulates membrane permeability, changes fluidity, and improves the stability of bilayer membranes in the presence of biological fluids.24 CHOL impacts the mechanical properties of vesicular membranes by enhancing their mechanical resilience, modifying membrane elasticity, and enhancing the lipid packing density through the effects of “ordering and condensing”.25 The inclusion of CHOL in the liposome results in a bilayer membrane that is less permeable to water, water-soluble molecules, small molecules, and ions. This property helps prevent interactions with plasma proteins, contributing to enhanced stability in biological fluids. So, the CHOL was used to increase the stability of the bilayer membrane.26 The filtration method using a 0.22 µm membrane filter was selected for sterilization purposes, as the liposomal formulations are thermolabile, may hydrolyze at the higher temperature of dry heat and moist heat sterilization, possibility of peroxidation & hydrolysis of unsaturated lipids during gamma irradiation. The formulations were kept protected from light and stored properly to avoid oxidation and photodegradation.
The Zavg and PDI of nanoformulations influence the endocytosis-dependent cellular uptake.27 The Zavg increased with the EL and CHOL concentrations (Table 2). The findings align with prior reports indicating that an increase in vesicle size corresponds to an increase in EL or CHOL.24,28-30 EL, as a primary component of the liposomal vesicular bilayer, leads to the formation of larger liposomal vesicles at higher concentrations. Similarly, CHOL, also a constituent of the liposomal bilayer, contributes to an elevation in Zavg with an increase in concentration.
The PDI value provides the homogeneity of Zavg distribution.18 Liposome and nanoliposome formulations, with a PDI of 0.3 and below, indicate a homogenous dispersion of phospholipid vesicles and are well accepted for drug delivery.27 The observed PDI values ranging from 0.253 ± 0.002 to 0.273 ± 0.005 suggest a narrow size distribution and homogeneous nature of the prepared liposomes.18,27
ζ values are crucial for maintaining colloidal dispersion stability through repulsive forces between particles.31 The ζ values for all formulations ranged from -17.8 ± 0.831 to -30.0 ± 0.29 mV (Table 2, Figure 2). With increasing the levels of CHOL after 10 mg, the zeta potential values decreased, indicating reduced surface charge, consistent with previous findings.25,32 Higher ζ values indicate increased repulsive forces between vesicles, enhancing colloidal stability. This elevated ζ helps prevent flocculation, coagulation, and sedimentation, contributing to prolonged stability.
The obtained pH value of liposome was found to be acidic (Table 2). For intravenous administration, the pH of the liposomes can be adjusted using a suitable buffer solution as the dispersion medium. The RI values of the liposomes (Table 2) were observed to closely approximate the RI of water (1.333), suggesting the transparency of the formulations. Further, the RI of three independent samples from liposomes were found to be closer to each other, indicating the isotropic nature of the system.
The entrapment efficiency was found to be increased with increasing the amounts of EL and CHOL. An increase in % EE at elevated EL levels can be attributed to the thorough distribution of the drug within the liposomal bilayer. Lipophilicity plays a crucial role in entrapping the lipophilic PLFEE in the lipid phase. Moreover, at higher EL concentrations, the Zavg increases, creating a larger space for the accumulation of the maximum quantity of the drug, resulting in a higher % EE. This finding aligns with previous reports indicating an increase in % EE of curcumin and doxorubicin with increased phospholipid concentrations.30 Conversely, the % LC was found to decrease with increasing EL and CHOL amounts, as the extract amount remains constant in all formulations.
As observed in the HRSEM photomicrographs (Figure 3a), the formulated liposomal vesicles exhibited a spherical shape and displayed uniformity in Zavg, featuring distinct and easily recognizable boundaries. However, some of the liposome vesicles are found to be irregular in shape, which might be due to the shrinkage of vesicles during drying for HRSEM analysis. The average vesicular size obtained from the size distribution graph (Figure 3b) was found to be as per the hydrodynamic Zavg obtained by dynamic light scattering (DLS). The 2D and 3D SPM photomicrographs (Figure 3c and Figure 3d) revealed the nanometric vesicle size, which is in accordance with the DLS and HRSEM results. The vesicular sizes obtained from the HRTEM photomicrograph (Figure 4a) are in accordance with the results obtained by DLS, HRSEM, and SPM. The diffused SAED pattern (Figure 4b) implied the amorphous nature of the PLFEE-loaded liposomes.33
The study of the drug-excipient interaction is important as it can affect the chemical integrity of the drug, influencing the safety and effectiveness of the prepared nanoformulations.34 FTIR, being a non-destructive vibrational spectroscopic tool, can offer valuable insights into chemical composition and interactions.35 All the vibrational peaks of PLFEE, EL, and CHOL were found to be preserved in the physical mixture (Figure 5a), which eradicates the possibility of incompatibility among them. All the important peaks of PLFEE are found to be present in the PLFEE-loaded LF3 without any shifting, which eliminates the possible interaction among the extract and excipients used in the liposomal formulation. However, the peaks of the freeze-dried liposomal formulation showed a decreased intensity due to the entrapment and dilution effect of the formulation excipients.
HPTLC fingerprinting was used as a qualitative analysis to identify the existence of the chemical groups of the standardized extract in the liposomal formulation. Any disappearance of chromatographic bands or shifting of the retardation factor of the phytochemicals in the formulation represents possible drug-excipient incompatibility in the developed formulation. The HPTLC fingerprints observed on the developed plates (Figure 5b) revealed the existence of chromatographic bands corresponding to pure piperine in both the standardized ethanolic extract and the formulation, representing the retention of its chemical integrity. Remarkably, all the chromatographic bands of the standardized PLFEE were found in the PLFEE-loaded LF3, affirming the maintenance of the chemical integrity of the extract in the formulation and the absence of drug-excipient incompatibility.
The liposome exhibited a biphasic release profile (Figure 6a), where the early rapid release is due to the release of surface-associated drugs, followed by sustained release attributed to the release of the vesicle-entrapped drug. The result is in accordance with the reported literature showing a biphasic release pattern of nanoliposomes.36,37 The limited accommodation in the liposomal bilayer results in the surface deposition of the free drug, which reaches initially. The sustained release profile from the liposome is linked to vesicular entrapment, acting as a drug reservoir and providing advantages over conventional dosage forms. The release profile of the liposome best conformed to Higuchi diffusion kinetics (Table 3). The major driving force for the diffusion-controlled release aligns with previous reports on liposomes.38 The release exponent (n = 0.598) from the Korsmeyer Peppas model represented anomalous diffusion.
The cytotoxicity by PLFEE-loaded liposome against B16F10 melanoma cell lines was found to be significantly improved (P < 0.05) than plain PLFEE (Figure 6b). The improved cytotoxicity of liposome is attributed to its nanovesicular structure, improved cell permeation, and greater partitioning capacity into the cells. The insignificant cytotoxicity of PLFEE and PLFEE-loaded liposomes towards healthy HEK293 cells (Figure 6c) represents its selective cytotoxicity without affecting the normal healthy cells. In a recent study, enhanced therapeutic efficacy of piperlongumine-loaded liposome has been reported for the treatment of cervical carcinoma compared to plain piperlongumine. Piperlongumine is one of the bioactive alkaloidal phytocomponents of P. longum L. However, its clinical outcomes are limited due to its hydrophobicity, low bioavailability, and rapid degradation. Piperlongumine-loaded nanoliposome was developed by thin-film hydration method using a quality-by-design approach. The liposome demonstrated improved cytotoxicity, reduced cell proliferation, decreased cell viability, improved nuclear condensation, inhibition of cell migration, reduction in mitochondrial membrane potential, increased reactive oxygen species level, and promoted more apoptosis in human cervical cancer cell lines (SiHa and HeLa) compared to unformulated piperlongumine. The nanoliposome demonstrated a potential therapeutic option for the treatment of cervical cancer.39
The stability study results (Table 4) for PLFEE-loaded liposomes revealed the maintenance of colloidal dispersion stability, physical stability, and chemical stability under refrigerated conditions for up to 3 months. Additionally, stability was demonstrated during freeze-thaw cycles and lyophilization, with no significant alterations observed in the stability-indicating parameters.
Conclusion
Cancer cells use various processes for their uncontrolled abnormal proliferation, invasion, angiogenesis, and metastasis. Therefore, the utilization of a multicomponent-based standardized herbal extract that targets multiple molecular pathways through diverse mechanisms is logical. Thin-film hydration method was chosen for the preparation of a standardized extract-loaded liposomal nanoformulation. The resulting liposomes were found to be homogeneous, amorphous, transparent, exhibited nano vesicular size, spherical morphology, excellent % EE, % LC, sustained drug release, drug-excipient compatibility, and remarkable stability, demonstrating selective cytotoxicity melanoma cells. The current study on the liposomal formulation containing the standardized ethanolic extract of P. longum L. holds promise as a potential alternative chemotherapeutic candidate for melanoma treatment. However, comprehensive in-vivo investigations in pre-clinical models and clinical trials are necessary to demonstrate its therapeutic potential for melanoma therapy.
Competing Interests
The authors state no conflict of interest.
Consent for Publication
Not applicable.
Data Availability Statement
All the data are included in this manuscript.
Ethical Approval
Not applicable.
Supplementary Files
Supplementary file 1. Liposomal Phytoformulation of Piper longum L. for Melanoma Therapy
(pdf)
Acknowledgements
The authors are extremely grateful to the Department of Pharmaceutical Engineering & Technology, IIT (BHU), Varanasi for providing infrastructure. The financial assistance provided as a scholarship to Himani, D Mohapatra, S Shreya, G G Naik, S D Malang, P Kathait, and Shambhavi by the Ministry of Human Resource Development (MHRD), Government of India, is greatly acknowledged. The fellowship provided to S Biswal by the Department of Biotechnology (DBT)-twinning, Govt. of India and the fellowship provided to P K Patel by the Council of Science and Technology, Uttar Pradesh, India, is highly acknowledged.
References
- Mohapatra D, Kumar DN, Shreya S, Panigrahi D, Agrawal AK, Sahu AN. Quality-by-design-based development of ultradeformable nanovesicular transgelosome of standardized Piper longum extract for melanoma. Nanomedicine (Lond) 2023; 18(14):963-85. doi: 10.2217/nnm-2023-0069 [Crossref] [ Google Scholar]
- Mohapatra D, Kumar DN, Shreya S, Pandey V, Dubey PK, Agrawal AK. Quality by design-based development and optimization of fourth-generation ternary solid dispersion of standardized Piper longum extract for melanoma therapy. Drug Deliv Transl Res 2023; 13(12):3094-131. doi: 10.1007/s13346-023-01375-y [Crossref] [ Google Scholar]
- Cancer Stat Facts: Melanoma of the Skin. Available from: https://seer.cancer.gov/statfacts/html/melan.html.
- Li J, Zhang Y, Tao J. Targeted nanoparticles for drug delivery to melanoma: from bench to bedside. In: Hamblin MR, Avci P, Prow TW, eds. Nanoscience in Dermatology. Boston: Academic Press; 2016. p. 203-15. doi: 10.1016/b978-0-12-802926-8.00016-1.
- Li J, Wang Y, Liang R, An X, Wang K, Shen G. Recent advances in targeted nanoparticles drug delivery to melanoma. Nanomedicine 2015; 11(3):769-94. doi: 10.1016/j.nano.2014.11.006 [Crossref] [ Google Scholar]
- Kratz F, Senter P, Steinhagen H. Drug Delivery in Oncology: From Basic Research to Cancer Therapy. John Wiley & Sons; 2013.
- Shreya S, Kasote D, Mohapatra D, Naik GG, Guru SK, Sreenivasulu N. Chemometric-based analysis of metabolomics studies of bioactive fractions of Pleurotusosteratus and their correlation with in vitro anti-cancer activity. Appl Biochem Biotechnol 2023; 195(7):4602-16. doi: 10.1007/s12010-023-04325-z [Crossref] [ Google Scholar]
- Albuquerque KR, Pacheco NM, Del Rosario Loyo Casao T, de Melo F, Novaes RD, Gonçalves RV. Applicability of plant extracts in preclinical studies of melanoma: a systematic review. Mediators Inflamm 2018; 2018:6797924. doi: 10.1155/2018/6797924 [Crossref] [ Google Scholar]
- Kinjo J, Nakano D, Fujioka T, Okabe H. Screening of promising chemotherapeutic candidates from plants extracts. J Nat Med 2016; 70(3):335-60. doi: 10.1007/s11418-016-0992-2 [Crossref] [ Google Scholar]
- Pradeep CR, Kuttan G. Piperine is a potent inhibitor of nuclear factor-kappaB (NF-kappaB), c-Fos, CREB, ATF-2 and proinflammatory cytokine gene expression in B16F-10 melanoma cells. Int Immunopharmacol 2004; 4(14):1795-803. doi: 10.1016/j.intimp.2004.08.005 [Crossref] [ Google Scholar]
- Fofaria NM, Kim SH, Srivastava SK. Piperine causes G1 phase cell cycle arrest and apoptosis in melanoma cells through checkpoint kinase-1 activation. PLoS One 2014; 9(5):e94298. doi: 10.1371/journal.pone.0094298 [Crossref] [ Google Scholar]
- Pradeep CR, Kuttan G. Effect of piperine on the inhibition of lung metastasis induced B16F-10 melanoma cells in mice. Clin Exp Metastasis 2002; 19(8):703-8. doi: 10.1023/a:1021398601388 [Crossref] [ Google Scholar]
- Song X, Gao T, Lei Q, Zhang L, Yao Y, Xiong J. Piperlongumine induces apoptosis in human melanoma cells via reactive oxygen species mediated mitochondria disruption. Nutr Cancer 2018; 70(3):502-11. doi: 10.1080/01635581.2018.1445769 [Crossref] [ Google Scholar]
- Brożyna AA, Jóźwicki W, Carlson JA, Slominski AT. Melanogenesis affects overall and disease-free survival in patients with stage III and IV melanoma. Hum Pathol 2013; 44(10):2071-4. doi: 10.1016/j.humpath.2013.02.022 [Crossref] [ Google Scholar]
- Sunila ES, Kuttan G. Piper longum inhibits VEGF and proinflammatory cytokines and tumor-induced angiogenesis in C57BL/6 mice. Int Immunopharmacol 2006; 6(5):733-41. doi: 10.1016/j.intimp.2005.10.013 [Crossref] [ Google Scholar]
- ICH Harmonised Tripartite Guideline. Validation of Analytical Procedures: Text and Methodology Q2(R1). Geneva, Switzerland: International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use; 2005. Available from: https://somatek.com/wp-content/uploads/2014/06/sk140605h.pdf.
- Varona S, Martín Á, Cocero MJ. Liposomal incorporation of lavandin essential oil by a thin-film hydration method and by particles from gas-saturated solutions. Ind Eng Chem Res 2011; 50(4):2088-97. doi: 10.1021/ie102016r [Crossref] [ Google Scholar]
- Yoon HY, Chang IH, Goo YT, Kim CH, Kang TH, Kim SY. Intravesical delivery of rapamycin via folate-modified liposomes dispersed in thermo-reversible hydrogel. Int J Nanomedicine 2019; 14:6249-68. doi: 10.2147/ijn.S216432 [Crossref] [ Google Scholar]
- Elmowafy M, Viitala T, Ibrahim HM, Abu-Elyazid SK, Samy A, Kassem A. Silymarin loaded liposomes for hepatic targeting: in vitro evaluation and HepG2 drug uptake. Eur J Pharm Sci 2013; 50(2):161-71. doi: 10.1016/j.ejps.2013.06.012 [Crossref] [ Google Scholar]
- Hamrapurkar PD, Jadhav K, Zine S. Quantitative estimation of piperine in Piper nigrum and Piper longum using high performance thin layer chromatography. J Appl Pharm Sci 2011; 1(3):117-20. [ Google Scholar]
- Pramod K, Suneesh CV, Shanavas S, Ansari SH, Ali J. Unveiling the compatibility of eugenol with formulation excipients by systematic drug-excipient compatibility studies. J Anal Sci Technol 2015; 6(1):34. doi: 10.1186/s40543-015-0073-2 [Crossref] [ Google Scholar]
- Perez-Ruiz AG, Ganem A, Olivares-Corichi IM, García-Sánchez JR. Lecithin-chitosan-TPGS nanoparticles as nanocarriers of (-)-epicatechin enhanced its anticancer activity in breast cancer cells. RSC Adv 2018; 8(61):34773-82. doi: 10.1039/c8ra06327c [Crossref] [ Google Scholar]
- Paradkar MM, Irudayaraj J. Determination of cholesterol in dairy products using infrared techniques: 1 FTIR spectroscopy. Int J Dairy Technol 2002; 55(3):127-32. doi: 10.1046/j.1471-0307.2002.00044.x [Crossref] [ Google Scholar]
- Nsairat H, Khater D, Sayed U, Odeh F, Al Bawab A, Alshaer W. Liposomes: structure, composition, types, and clinical applications. Heliyon 2022; 8(5):e09394. doi: 10.1016/j.heliyon.2022.e09394 [Crossref] [ Google Scholar]
- Magarkar A, Dhawan V, Kallinteri P, Viitala T, Elmowafy M, Róg T. Cholesterol level affects surface charge of lipid membranes in saline solution. Sci Rep 2014; 4:5005. doi: 10.1038/srep05005 [Crossref] [ Google Scholar]
- Jovanović AA, Balanč BD, Ota A, Ahlin Grabnar P, Djordjević VB, Šavikin KP. Comparative effects of cholesterol and β‐sitosterol on the liposome membrane characteristics. Eur J Lipid Sci Technol 2018; 120(9):1800039. doi: 10.1002/ejlt.201800039 [Crossref] [ Google Scholar]
- Danaei M, Dehghankhold M, Ataei S, Hasanzadeh Davarani F, Javanmard R, Dokhani A. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018; 10(2):57. doi: 10.3390/pharmaceutics10020057 [Crossref] [ Google Scholar]
- Trivedi R, Kotgale NR, Taksande J, Wadher K, Awandekar NB, Umekar M. Influence of egg lecithin composition on physicochemical characteristics of pramipexole liposomes. Int J Res Pharm Sci 2017; 8(1):6-15. [ Google Scholar]
- Kaddah S, Khreich N, Kaddah F, Charcosset C, Greige-Gerges H. Cholesterol modulates the liposome membrane fluidity and permeability for a hydrophilic molecule. Food Chem Toxicol 2018; 113:40-8. doi: 10.1016/j.fct.2018.01.017 [Crossref] [ Google Scholar]
- Tefas LR, Sylvester B, Tomuta I, Sesarman A, Licarete E, Banciu M. Development of antiproliferative long-circulating liposomes co-encapsulating doxorubicin and curcumin, through the use of a quality-by-design approach. Drug Des Devel Ther 2017; 11:1605-21. doi: 10.2147/dddt.S129008 [Crossref] [ Google Scholar]
- Vijayakumar MR, Kosuru R, Vuddanda PR, Singh SK, Singh S. Trans resveratrol loaded DSPE PEG 2000 coated liposomes: an evidence for prolonged systemic circulation and passive brain targeting. J Drug Deliv Sci Technol 2016; 33:125-35. doi: 10.1016/j.jddst.2016.02.009 [Crossref] [ Google Scholar]
- Banerjee KK, Maity P, Das S, Karmakar S. Effect of cholesterol on the ion-membrane interaction: zeta potential and dynamic light scattering study. Chem Phys Lipids 2023; 254:105307. doi: 10.1016/j.chemphyslip.2023.105307 [Crossref] [ Google Scholar]
- Erceg I, Kontrec J, Strasser V, Selmani A, Domazet Jurašin D, Ćurlin M. Precipitation of calcium phosphates and calcium carbonates in the presence of differently charged liposomes. Minerals 2022; 12(2):208. doi: 10.3390/min12020208 [Crossref] [ Google Scholar]
- Fredenberg S, Wahlgren M, Reslow M, Axelsson A. The mechanisms of drug release in poly(lactic-co-glycolic acid)-based drug delivery systems--a review. Int J Pharm 2011; 415(1-2):34-52. doi: 10.1016/j.ijpharm.2011.05.049 [Crossref] [ Google Scholar]
- Pachauri M, Gupta ED, Ghosh PC. Piperine loaded PEG-PLGA nanoparticles: Preparation, characterization and targeted delivery for adjuvant breast cancer chemotherapy. J Drug Deliv Sci Technol 2015; 29:269-82. doi: 10.1016/j.jddst.2015.08.009 [Crossref] [ Google Scholar]
- Sharma R, Yadav V, Katari O, Jain S. Hyaluronic acid functionalized liposomes for co-delivery of paclitaxel and ursolic acid for enhanced efficacy against triple negative breast cancer. J Drug Deliv Sci Technol 2025; 104:106451. doi: 10.1016/j.jddst.2024.106451 [Crossref] [ Google Scholar]
- Srinath P, Vyas SP, Diwan PV. Preparation and pharmacodynamic evaluation of liposomes of indomethacin. Drug Dev Ind Pharm 2000; 26(3):313-21. doi: 10.1081/ddc-100100359 [Crossref] [ Google Scholar]
- Gibis M, Ruedt C, Weiss J. In vitro release of grape-seed polyphenols encapsulated from uncoated and chitosan-coated liposomes. Food Res Int 2016; 88(Pt A):105-13. doi: 10.1016/j.foodres.2016.02.010 [Crossref] [ Google Scholar]
- Parveen S, Kumar S, Pal S, Yadav NP, Rajawat J, Banerjee M. Enhanced therapeutic efficacy of Piperlongumine for cancer treatment using nano-liposomes mediated delivery. Int J Pharm 2023; 643:123212. doi: 10.1016/j.ijpharm.2023.123212 [Crossref] [ Google Scholar]