Pharmaceutical Sciences. 2025;31(3):294-303.
doi: 10.34172/PS.025.40914
Research Article
Phytosterol Glycosides from Olax subscorpioidea Oliv. Exhibit Cytotoxic Effects in In Vitro and In Silico Studies
Yemi A. Adekunle Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, 1, 2, 3, * 
Babatunde B. Samuel Conceptualization, Formal analysis, Supervision, 1 
Lutfun Nahar Funding acquisition, Supervision, Writing – review & editing, 4, * 
Amos A. Fatokun Writing – review & editing, 2 
Satyajit D. Sarker Supervision, Validation, Writing – review & editing, 2 
Author information:
1Department of Pharmaceutical Chemistry, Faculty of Pharmacy, University of Ibadan, Nigeria
2Centre for Natural Products Discovery, School of Pharmacy and Biomolecular Sciences, Faculty of Science, Liverpool John Moores University, Byrom Street, Liverpool L3 3AF, United Kingdom
3Department of Pharmaceutical and Medicinal Chemistry, College of Pharmacy, Afe Babalola University, Ado-Ekiti, Nigeria
4Laboratory of Growth Regulators, Palacký University and Institute of Experimental Botany, The Czech Academy of Sciences, Šlechtitelů 27, 78371 Olomouc, Czech Republic
Abstract
Background:
Olax subscorpioidea is traditionally used to treat arthritis, cancer, diabetes, neurodegenerative disorders, and oxidative stress. This study carried out chromatographic isolation, cytotoxicity, and molecular docking studies of bioactive compounds from O. subscorpioidea.
Methods:
The root of O. subscorpioidea was extracted with methanol using the Soxhlet extraction method. The extract was partitioned into n-hexane, dichloromethane (DCM), and methanol/water. The DCM fraction was subjected to column chromatography. Bioactive compounds were isolated and their chemical structures were established by one-dimensional (1D) and two-dimensional (2D) nuclear magnetic resonance (NMR) spectroscopy, and by comparing their NMR data with those previously reported in the literature. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was used to evaluate the cytotoxic activity of these compounds against three human cancer cell lines: breast (MCF-7), cervical (HeLa), and colorectal (Caco-2) cell lines. Molecular docking was used to gain insights into the favourable binding conformations and energies of the compounds when interacting with ten selected cancer-related protein targets.
Results:
The phytochemical investigation of the extract of O. subscorpioidea afforded two sterol glycosides, stigmast-5,22-dien-3-O-β-D-glucoside (1a) and sitosterol-3-O-β-D-glucoside (1b), as a mixture. The compounds were found to be active against HeLa (IC50: 37.0±4.51 µg/mL) and MCF-7 (137.07±19.43 µg/mL) cell lines. The compounds showed strong interactions with the colchicine-binding site on the β-subunit of tubulin protein, epidermal growth factor receptor kinase domain, poly(ADP-ribose) polymerase-1, and 17β-hydroxysteroid dehydrogenase type 1 (binding energies: –10.3 and –10.0 kcal/mol; –9.3 and –9.3 kcal/mol; –9.2 and –9.2 kcal/mol; and –9.3 and –9.1 kcal/mol, respectively). Stigmast-5,22-dien-3-O-β-D-glucoside was consistently ranked higher in some of the proteins tested. The compounds stabilised in the binding sites of the proteins via hydrogen bonds and hydrophobic interactions.
Conclusion:
To the best of our knowledge, this is the first report on the isolation of these compounds from this plant. The cytotoxic effects of O. subscorpioidea root extract could be partly attributed to stigmast-5,22-dien-3-O-β-D-glucoside and sitosterol-3-O-β-D-glucoside.
Keywords: Olax subscorpioidea, Cancer, NMR, Molecular docking, Stigmast-5,22-dien-3-O-β-D-glucoside, Sitosterol-3-O-β-D-glucoside
Copyright and License Information
© 2025 The Author(s).
This is an open access article and applies the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
This study was supported by the following grants: a Commonwealth Split-Site PhD Fellowship awarded to Yemi A. Adekunle (Ref.: NGCN-2021-184); the European Regional Development Fund—Project ENOCH (No. CZ.02.1.01/0.0/0.0/16_019/0000868); and Czech Agency Grants (Projects 23-05474S and 23-05389S) awarded to Lutfun Nahar.
Introduction
Plants have provided medicine for humans for many centuries.1,2 A large percentage of the world’s population, especially in developing countries, depend on medicinal plants as sources of medicines.3 Secondary metabolites have been recognised as the healing principles in plants. Cancer is a leading cause of death globally.4 It kills about 10 million people every year across the globe.5 Approximately 70% of cancer-related deaths occur in low- and middle-income countries.6 Many cancer drugs have been developed, the majority of which have serious adverse effects.1,7 Therefore, there is a need for anticancer drugs that are more tolerable, effective, and less toxic.
Olax subscorpioidea Oliv. belongs to the Olacaceae family that comprises shrubs, trees and woody dicotyledonous plants with significant economic and medicinal values. This family includes a range of secondary metabolites, such as flavonoids, triterpenoids, and secoiridoids, each exhibiting diverse biological activities.8-10 The plants, distributed within about 25 genera, are widely distributed in temperate and tropical regions.9
Olax subscorpioidea is a shrub-like tree that reaches a height of approximately 10 metres.11 The plant is traditionally used to treat arthritis, cancer, depression, diabetes mellitus, neurodegenerative disorders, oxidative stress, and swelling.12,13 Ethnobotanical surveys showed that O. subscorpioidea is an important ingredient in traditional cancer remedies in Nigeria.14-16 We recently reported cytotoxic triterpenoid saponins from O. subscorpioidea.17
The preliminary cytotoxic analysis of the methanol root extract of O. subscorpioidea showed antiproliferative activity against human rhabdomyosarcoma (RD) and breast cancer (MCF-7) cell lines.18 The current study, therefore, aimed at isolating and characterising bioactive compounds from O. subscorpioidea root extract. The potential of these compounds as inhibitors of the growth of human cancer cell lines was evaluated by the MTT assay. Molecular docking and ADMET (absorption, distribution, metabolism, excretion, toxicity) studies were also conducted.
Methods
General experimental procedures
The infrared spectroscopy (IR) spectrum was obtained using a Cary 630 FTIR spectrophotometer (Agilent Technologies, USA). 1D and 2D nuclear magnetic resonance (NMR) data were obtained on a Bruker Avance III NMR spectrometer at 600 MHz (Bruker, USA). The sample was dissolved in dimethyl sulfoxide (DMSO-d6) and the solvent residue served as the internal standard. Chemical shifts (δ) were measured in parts per million (ppm), and coupling constants were measured in hertz (Hz). High-performance liquid chromatography (HPLC) grade solvents from Fisher Scientific (Loughborough, United Kingdom) were used. Cell culture materials include Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, UK), L-glutamine 200 mM (100X) (Gibco, UK), antibiotic-antimycotic (100X) (Gibco, UK), TrypLE Express (1X) (Thermo Fisher Scientific, UK), Fetal bovine serum (Sigma-Aldrich, UK), Phosphate buffered-saline (Thermo Fisher Scientific, UK), and MTT (Sigma-Aldrich, UK). Vinblastine from Tocris (UK) was used as a reference drug. Absorbance measurements were performed on a Tecan Spark 10M multimode microplate reader (Switzerland).
Plant collection
The root of O. subscorpioidea was collected in April 2021 from the University of Ibadan botanical garden, Ibadan, Nigeria. The collection and identification of the plant were performed by the garden curator, Mr Michael Owolabi. A sample was deposited at the Forest Herbarium, Ibadan (FHI) with voucher number FHI:113182. The root material was cleaned, peeled, and dried in the lab. After that, it was ground into powder.
Extraction and fractionation
Approximately 600 g of powdered O. subscorpioidea root material was extracted with methanol using the Soxhlet extraction method. The extract was concentrated in vacuo using a rotary evaporator at a temperature below 40 °C (Heidolph HB Digital, Germany). The resulting extract was dried at room temperature in the laboratory and stored for later use. About 63.7 g of extract was obtained. The extract was further partitioned into n-hexane, dichloromethane (DCM), and water/methanol. Approximately 40 g of the extract was dissolved in water/methanol (20/80%) in a separating funnel and partitioned with an equal volume of n-hexane. The hexane fraction was obtained and concentrated. The extraction was done three times. Into the aqueous fraction, an equal volume of DCM was added (extraction was carried out three times) and the fraction was concentrated. The aqueous fraction was also obtained and concentrated.
Isolation of bioactive compounds
The isolation of compounds was carried out on an open glass chromatographic column packed with 60-120 mesh silica gel. The DCM fraction, 5.48 g, was re-suspended and loaded on the chromatography column (75 cm × 3.5 cm). The sample was eluted with n-hexane/ethyl acetate 70%:30% to 100% ethyl acetate, followed by ethyl acetate/methanol, 90%:10%; 80%:20%; 70%:30%; 50%:50%; 30%:70%; and 0%:100%. A total of 64 fractions were obtained (approximately, 30 mL each), which were pooled into 5 (F1–F5) according to their retention factor (Rf) pattern (F254 silica gel 60 TLC plate, 70/20/10% n-hexane/ethyl acetate/methanol). Fraction F4 was further purified by crystallisation to obtain a white amorphous powder. The substance contained a mixture of two compounds assigned as 1 (9.8 mg). The compound showed a retention factor (Rf) of 0.6 in ethyl acetate/methanol 9.5/0.5%. Compound 1 was visualised under an ultraviolet (UV) lamp (UVITEC, Cambridge). The developed TLC plate (Figure S1) was detected by being sprayed with conc. H2SO4, and then left in the oven at 110℃ for about 10 min (Oven DHG-9053A, Ocean Med, England).
Cytotoxic analysis
Cells and cell culture
Human Caco-2, HeLa, and MCF-7 cancer cell lines were originally obtained from the American Type Culture Collection (ATCC) or the European Collection of Authenticated Cell Cultures (ECACC). As previously described,17 the cells were cultured in DMEM supplemented with 10% Foetal Bovine Serum (FBS), 1% 2 mM L-glutamine, and 1% penicillin-streptomycin-amphotericin B solution. They were maintained in a 37 °C incubator with a 5% CO2 humidified atmosphere. Cells were passaged approximately twice weekly. Before the treatment, cells were seeded into 96-well microplates and incubated for 24 hours. Cell visualization (at × 10 magnification) was performed using an Olympus CKX41 inverted microscope.
Cell treatments and cell viability assessment with the MTT assay
Stock solutions were prepared in DMSO, and all further dilutions were made in full growth medium. The highest concentrations used contained no more than 0.1% DMSO, which was non-toxic to the cells. Each well containing approximately 7,500 cells, prepared as previously reported,17 was treated with the sample (concentrations: 1, 10, 50, 100, and 200 µg/mL). Each experiment was performed in triplicate and repeated three independent times (n = 3). The effects on the viability of the three human cell lines of the isolated compounds were assessed using the in vitro MTT assay.19
Molecular docking study
Ligand, protein retrieval and preparation
The ligands used in this study were the structures of stigmast-5,22-dien-3-O-β-D-glucoside and sitosterol-3-O-β-D-glucoside isolated from the methanol root extract of O. subscorpioidea. Their structures were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The 3D structures were prepared for docking using the UCSF Chimera tool (1.16).20 Gasteiger charges and hydrogen atoms were added.21 Energy minimisation of the structures was carried out at 100 steepest descent steps.
The 3D structures of the following proteins were retrieved from the Protein Data Bank (PDB) (https://www.rcsb.org/).22 Alpha-beta tubulin complexed with taxol (PDB ID: 1JFF),23 alpha-beta tubulin complexed with colchicine (PDB ID: 1SA0),24 caspase-3 (PDB ID: 1GFW),25 human 17 beta-hydroxysteroid dehydrogenase-1 (PDB ID: 1FDW),26 phosphoinositide 3-kinase (PDB ID: 1E8Z),27 poly (ADP-ribose) polymerase-1 (PDB ID: 5DS3),28 bromodomain-containing protein 4 (PDB ID: 7JKW),29 epidermal growth factor receptor tyrosine kinase domain (PDB ID: 1M17),30 human oestrogen receptor alpha (PDB ID: 3ERT),31 and polo-like kinase-1 (PDB ID: 3FC2).32 To prepare the proteins, they were individually uploaded to the workspace of Chimera. All ligands, small molecules, water, and other co-crystallised compounds were deleted. Hydrogen atoms were added to the proteins. In addition, partial charges were added by ANTECHAMBER. The structures were then minimised to 200 steepest descent steps at 10-minute intervals. The prepared proteins were saved in.pdb to be used for docking.33
Docking
A molecular docking study was carried out to obtain information about the various binding conformations and binding energies of the compounds when interacting with the selected proteins. A site-directed docking was performed for all proteins. First, they were uploaded to the GPU of the Python Prescription Virtual Screening Tool (PyRx). They were then made into macromolecules. The active site of each protein had been pre-determined as the residues within 5 Å region of the co-crystallised ligand in Chimera. The ligands were imported to the GPU of PyRx, converted to AutoDock ligand format (.pdbqt), and the grid box set, fitting the active site which was defined by the grid box coordinates as shown in Table 1. Docking was carried out with the AutoDock Vina of PyRx.34 BIOVIA Discovery Studio Visualiser was used to analyse the protein-ligand interactions.21
Table 1.
Binding site coordinates of some protein targets
|
Dimensions
|
17
β
HDS1 (PDB ID: 1FDW)
|
PARP-1 (PDB ID: 5DS3)
|
EGFR (PDB ID: 1M17)
|
αβ
-TUBULIN (PDB ID:1SA0)
|
| Centre_x (Å) |
46.2939 |
– 1.9486 |
24.6136 |
117.8476 |
| Centre_y (Å) |
7.0436 |
37.6889 |
–0.3664 |
89.7445 |
| Centre_z (Å) |
42.6282 |
13.7477 |
25.3674 |
6.5756 |
| Size_x (Å) |
23.8829 |
23.1186 |
28.0573 |
19.6719 |
| Size_y (Å) |
20.8675 |
18.1855 |
18.3221 |
25.6549 |
| Size_z (Å) |
22.8567 |
26.5048 |
21.7736 |
20.9588 |
ADMET study
Drug-likeness means how much a compound’s properties resemble those of existing drugs. The physicochemical parameters and ADME properties of the isolated compounds were evaluated using a freely available web tool, SwissADME (http://www.swissadme.ch/).35 Some of the parameters obtained include molecular weight, lipophilicity, hydrogen bond acceptors and donors, topological polar surface area (TPSA), enzyme inhibition, and synthetic route accessibility.
Statistical analysis
Each experiment was carried out three independent times (n = 3). Data is presented as mean of IC50 ± SEM (standard error of the mean). GraphPad Prism (9.4) was used to calculate the IC50. One-way ANOVA was used to determine the statistical significance of differences between means (P < 0.05 considered significant).
Results and Discussion
Cytotoxic effects of O. subscorpioidea fractions
Olax subscorpioidea is found in many countries across West and Central Africa and its traditional applications include its use to treat cancer, diabetes mellitus, neurodegenerative disorders, and swelling.12,13 An ethnobotanical study among people in Ogun State, Nigeria, discovered that a mixture containing O. subscorpioidea root, along with other plants including Alafia barteri leaf, Anthocleista djalonensis root, Calliandra portoricensis root, Clausena anisata stem bark, Macaranga barteri stem bark, Tephrosia vogelii stem bark, Triclisia subcordata leaf, Xylopia aethiopica, and potash, is used to treat breast cancer.15 A preliminary study of the cytotoxic activity of O. subscorpioidea revealed activity against MCF-7 and RD.18 The methanol root extract was further fractionated into n-hexane, DCM, and aqueous media. The DCM fraction showed the highest activity against the two cell lines with percentage cell inhibition of 80.08% for MCF-7 and 82.01% for RD. The activity was closely followed by that of the n-hexane fractionwith percentage cell inhibition of 78.42% for MCF-7 and 79.98% for RD. The aqueous fraction showed lower activity, having cell inhibition of 8.46% for MCF-7 and 40.87% for RD.18 The current study focused on isolating bioactive compounds from the DCM fraction and evaluating their cytotoxic effects against human cancer cell lines.
Structure determination
The DCM fraction yielded a white amorphous powder, 9.8 mg (Compound 1). Extensive NMR experiments revealed that the isolated compound was a mixture of two known sterol glycosides. In the 13C NMR experiment, the signals at δC 101.05 and 101.27, and 121.66 and 121.69 occurred as pairs each indicating a mixture of two similar compounds. The heteronuclear multiple bond correlation (HMBC) spectrum was used to verify the identity of the individual compounds in each mixture, which were identified as stigmasterol glucoside and β-sitosterol glucoside by detecting the distinct signals at positions C-22 and C-23.36 Using HMBC spectrum, the chemical shifts of stigmasterol glucoside at positions C-22 and C-23 were confirmed as 138.5 and 129.3 ppm, respectively, indicating the presence of an alkene group. In contrast, β-sitosterol glucoside showed chemical shift signals for C-22 and C-23 resonating at 33.8 and 25.9 ppm, respectively, suggesting an alkyl group (Table S1). Homonuclear correlation spectroscopy (1H-1H COSY), heteronuclear single quantum correlation (HSQC), and HMBC spectra helped confirm all 1H and 13C assignments (Figures 1a and 1b, and Figures S2–S7).
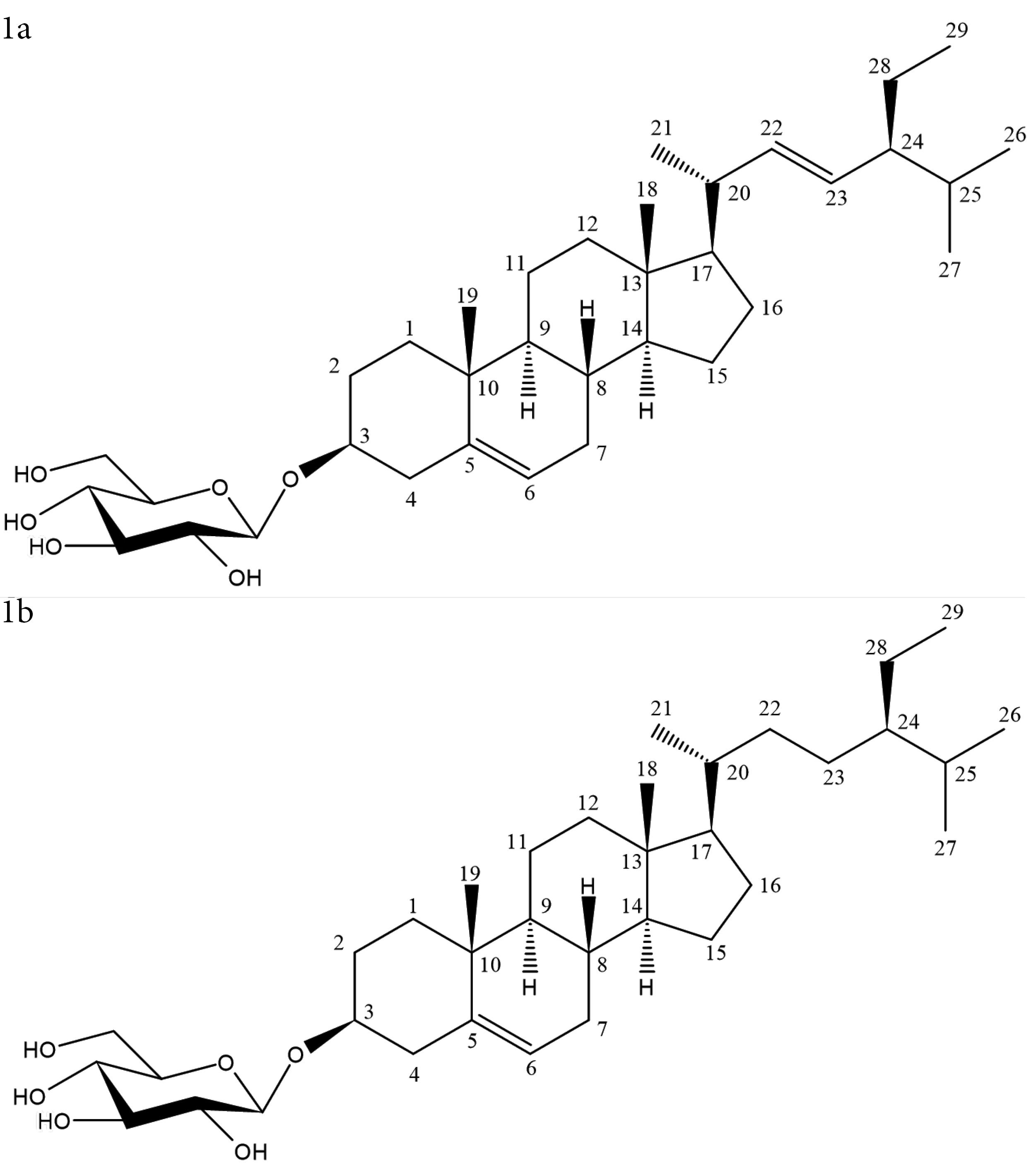
Figure 1.
Structure of compound 1a and 1b
.
Structure of compound 1a and 1b
Stigmast-5,22-dien-3-O-β-D-glucoside (1a), white amorphous powder. IR: 3360 cm-1, broad (OH stretch), 2910 cm-1 (CH stretch), and 1010 cm-1 (CO stretch). 1H NMR (DMSO, 600 MHz): δH 2.12, 2.35 (2H, C-1), 3.45 (m, 1H, C-3), 1.79 (2H, C-4), 5.33 (t, 1H, C-6), 1.91 (2H, C-7), 1.43 (1H, C-8), 0.99 m (1H, C-14), 1.79 (2H, C-16), 0.66 (s, 3H, C-18), 0.95 (s, 3H, C-19), 0.99 (d 6.48 Hz, 3H, C-21), 5.15 (m, 1H, C-22), 5.02 (m, 1H, C-23), 1.50 (1H, C-24), 1.34 (1H, C-25), 0.82 (3H, C-26), 0.78 (d 2.32 Hz, 3H, C-27), 0.77 (3H, C-29), 4.20 (d, 7.84 Hz, C-1’). 13C-DEPT-Q δC: 38.8 (C-1), 77.4 (C-3), 37.3 (C-4), 140.9 (C-5), 121.7 (C-6), 31.8 (C-7), 55.8 (C-17), 12.3 (C-18), 19.6 (C-19), 21.6 (C-21), 138.5 (C-22), 129.3 (C-23), 51.1 (C-24), 31.9 (C-25), 21.4 (C-26), 19.3 (C-27), 12.6 (C-29), 101.2 (C-1’) 73.9 (C-2’), 77.2 (C-3’), 70.5 (C-4’), 77.2 (C-5’), 61.5 (C-6’).37-40
Sitosterol-3-O-β-D-glucoside (1b), white amorphous powder. IR: 3360 cm-1, broad (OH stretch), 2910 cm-1 (CH stretch), and 1010 cm-1 (CO stretch). 1H NMR (DMSO, 600 MHz): δH 3.45 m (1H, C-3), 5.33 (t, 1H, C-6), 1.91 (2H, C-7), 0.99 m (1H, C-14), 1.38 (2H, C-15), 1.78 (2H, C-16), 1.13 (1H, C-17), 0.66 (s, 3H, C-18), 0.80 (s, 3H, C-19), 1.33 (1H, C-20), 0.95 (3H, C-21), 1.00, 1.29 (2H, C-22), 1.14 (2H, C-23), 0.91 (1H, C-24), 0.90 (d, 6.22 Hz C-27), 0.81 (3H, C-29), 4.20 (d, 7.84 Hz C-1’), 3.08 (1H, C-3’), 3.04 (1H, C-5’), 3.39, 3.63 (1H, C-6’). 13C-DEP-Q δC: 36.7 (C-1), 29.0 (C-2), 77.3 (C-3), 140.9 (C-5), 121.7 (C-6), 50.1 (C-9), 36.7 (C-10), 21.1 (C-11), 37.3 (C-12), 56.6 (C-14), 25.4 (C-15), 28.3 (C-16), 55.9 (C-17), 12.2 (C-18), 20.2 (C-19), 36.0 (C-20), 19.4 (C-21), 33.8 (C-22), 25.9 (C-23), 45.6 (C-24), 29.2 (C-25), 19.4 (C-26), 19.1 (C-27), 23.1 (C-28), 12.3 (C-29), 101.1 (C-1’), 73.9 (C-2’), 77.2 (C-3’), 70.5 (C-4’), 77.2 (C-5’), 61.5 (C-6’).38,41,42
The occurrence of β-sitosterol and stigmasterol or their glycosides together in the same plant is common. Similarly, the isolation of the two compounds or their glycosides together as a mixture is also frequent. Kamal et al36 isolated a mixture of β-sitosterol and stigmasterol from Vitex pinnata L. Ekhuemelo et al43 isolated a mixture of sitosterol, stigmasterol and cycloeucalenol from Erythrophleum suaveolens (Guill. & Perr.) Brenan. Here are a few other plants from which a mixture of the compounds was reported: Baccaurea macrocarpa Miq. Mull. Arg,44 Cassia sieberiana DC.,45 Premna herbacea Roxb.,46 and Ricinus communis L.47 Elfita et al48 isolated a mixture of their glycosides from Garcinia griffithii T. Anderson. These compounds also exhibit a variety of biological activities.
Cytotoxic effects of the isolated compounds
The cytotoxic potencies of the isolated compounds, as indicated by the IC50 values, are presented in Table 2. Compound 1, a mixture of stigmast-5,22-dien-3-O-β-D-glucoside (1a) and sitosterol-3-O-β-D-glucoside (1b), showed cytotoxicity against the HeLa and MCF-7 cell lines. Its most potent cytotoxicity was against the HeLa cell line, with an IC50 value of 37.0 ± 4.51 µM, although it also exhibited mild cytotoxicity against the MCF-7 cell line, with an IC50 value of 137.07 ± 19.43 µg/mL (Table 2). Ikpefan et al49 reported that stigmast-5,22-dien-3-O-β-D-glucoside was sensitive to MCF-7 and NCI-H460 cell lines with GI50 values of 40.83 ± 0.1 and 58.83 ± 11.2 µM, respectively, using the sulforhodamine B (SRB) cytotoxicity assay. Quradha et al42 reported that sitosterol-3-O-β-D-glucoside had moderate activity against the HeLa cell line, with a cell inhibition of 48%, and a weak effect on the 3T3 cell line (mouse fibroblast), with a cell inhibition of 39%. This compound isolated from the aerial part of Cynara cardunculus var. altilis was also shown to have an IC50 value of 53.6 µg/mL against the hepatocellular carcinoma (HepG2) cell line.50 A study investigating the effects of sitosterol-3-O-β-D-glucoside on HepG2 and SMMC-7721 cells, via the Wnt/β-catenin signalling pathway, found that it inhibited cell migration and invasion in HepG2 cells through this signalling pathway.51 Another study revealed that sitosterol-3-O-β-D-glucoside exhibited its cytotoxic effects through the induction of apoptosis and activation of caspase-3 and -9 in the hepatocellular (Huh7 and HepG2) cancer cell lines.52
Table 2.
Cytotoxic potencies (IC50) of compounds 1a and 1b from O. subscorpioidea against human cancer cell lines
|
IC50±SEM (µg/mL)
|
|
Compounds
|
HeLa
|
MCF-7
|
Caco-2
|
|
1
|
37.0 ± 4.51 |
137.07 ± 19.43 |
> 200 |
| Vincristine |
1.42 ± 0.40* |
0.54 ± 0.06* |
1.33 ± 0.05* |
* Activities expressed in µM.
Molecular docking study
Molecular docking is a crucial computational tool in predicting binding conformations and binding energies of small molecule drugs (ligands) to receptor proteins, which enables high-throughput virtual screening.53 In this study, ten cancer-related proteins were selected for the docking study. The compounds docked well with all the proteins. Stigmast-5,22-dien-3-O-β-D-glucoside consistently showed stronger interactions than sitosterol-3-O-β-D-glucoside with some of the proteins. The best interactions, with the least negative binding energies, were observed with alpha-beta tubulin complexed with colchicine (PDB ID: 1SA0). Compounds 1a and 1b showed binding energy of –10.3 and –10.0 kcal/mol, respectively. For compound 1a, this was followed by epidermal growth factor receptor tyrosine kinase domain (EGFR) (PDB ID: 1M17) and 17β-hydroxysteroid dehydrogenase-1 (17β-HSD1) (PDB ID: 1FDW) with a binding energy of –9.3 kcal/mol. Poly (ADP-ribose) polymerase-1 (PARP-1) (PDB ID: 5DS3) had a binding energy of –9.2 kcal/mol for compound 1a. For compound 1b, EGFR ranked second with a binding energy of –9.3 kcal/mol, while PARP-1 had a binding energy of –9.2 kcal/mol (Table 3). The two compounds showed the least binding interactions with bromodomain-containing protein 4 (BRD4) (PDB ID: 7JKW) and caspase-3 (PDB ID: 1GFW).
Table 3.
Binding affinities of isolated compounds against ten selected protein targets
|
Ligands
|
PubChem ID
|
Binding affinities (kcal/mol)
|
|
1JFF
|
1GFW
|
1FDW
|
1E8Z
|
5DS3
|
7JKW
|
1M17
|
1SA0
|
3ERT
|
3FC2
|
|
1a
|
6602508 |
–9.0 |
–7.8 |
–9.3 |
–8.3 |
–9.2 |
–7.7 |
–9.3 |
–10.3 |
–8.2 |
–8.8 |
|
1b
|
12309057 |
–7.9 |
–7.7 |
–9.1 |
–7.7 |
–9.2 |
–7.4 |
–9.3 |
–10.0 |
–8.0 |
–8.8 |
The compounds show similar conformations at the colchicine-binding site of alpha-beta tubulin (Figures 2A and 3E). αβ-Tubulin heterodimers are microtubule proteins that are crucial for the mitotic phase of cell division. Classical inhibitors of the β-tubulin subunit act by promoting or disrupting microtubule assemblage during mitosis by binding to the colchicine-, vinca-, or taxol-binding sites.54 The isolated compounds form a bridge between the α- and β-subunits of tubulin. The compounds were orientated in such a way that the polar glycosidic tail pointed toward the α-chain, while the non-polar head formed hydrophobic interactions with the β-chain. The compounds were observed to stabilise at the colchicine-binding pocket with crucial amino acid residues such as threonine (THR) 179 and cysteine (CYS) 241, the residues that have been implicated in the binding of colchicine-like compounds to the protein.54 Similarly, colchicine hydrogen bonds with the CYS241 residue of β-tubulin and shows hydrophobic interaction.55
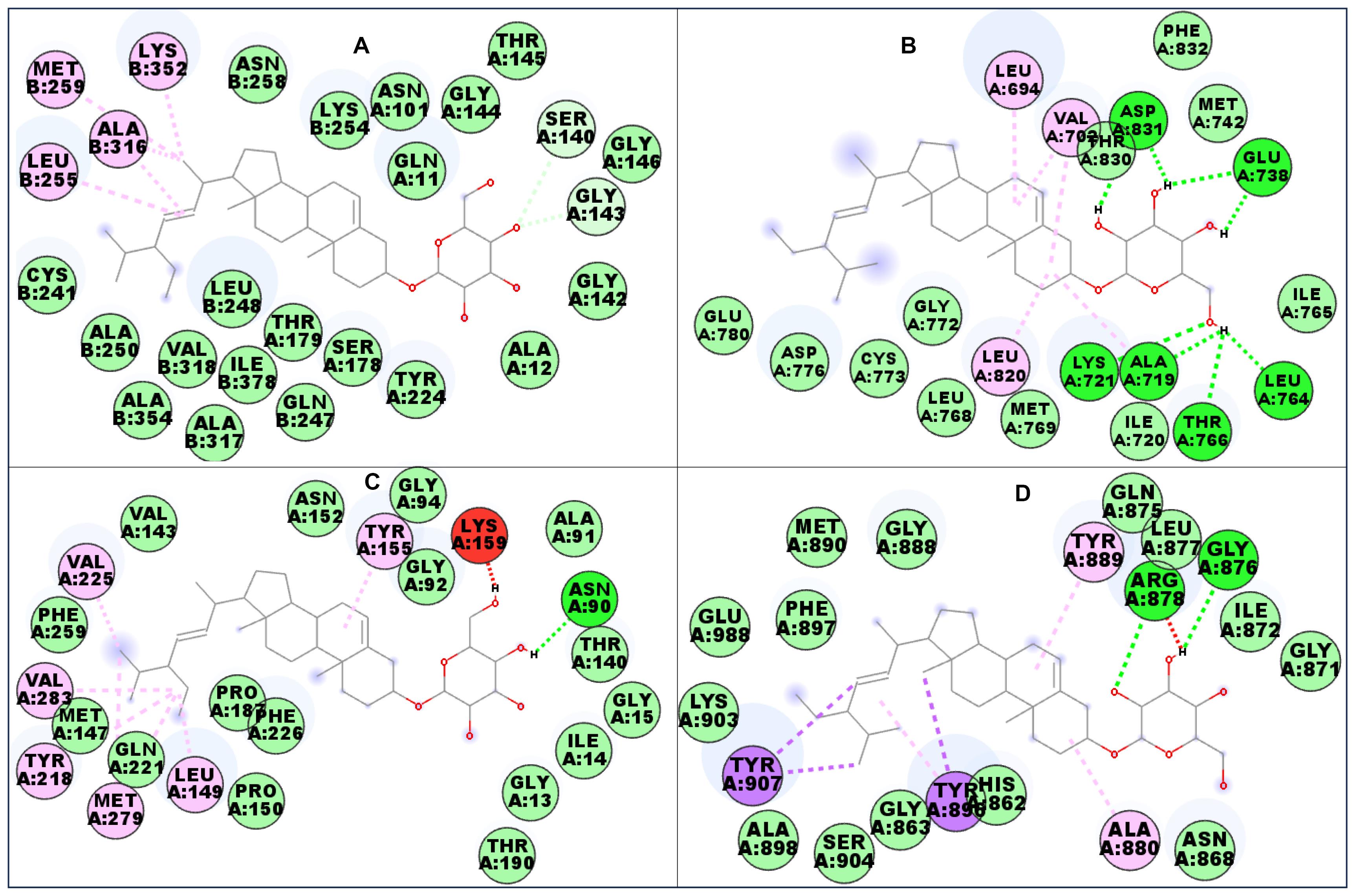
Figure 2.
Interacting residues of (A) Alpha-beta tubulin, (B) EGFR, (C) 17β-HSD1, and (D) PARP-1 with compound 1a. Green: Conventional hydrogen bonding. Lilac: Alkyl/pi-alkyl interaction. Blue: Carbon hydrogen bonding. Purple: Pi-sigma bonding. Light green: van der Waals forces
.
Interacting residues of (A) Alpha-beta tubulin, (B) EGFR, (C) 17β-HSD1, and (D) PARP-1 with compound 1a. Green: Conventional hydrogen bonding. Lilac: Alkyl/pi-alkyl interaction. Blue: Carbon hydrogen bonding. Purple: Pi-sigma bonding. Light green: van der Waals forces
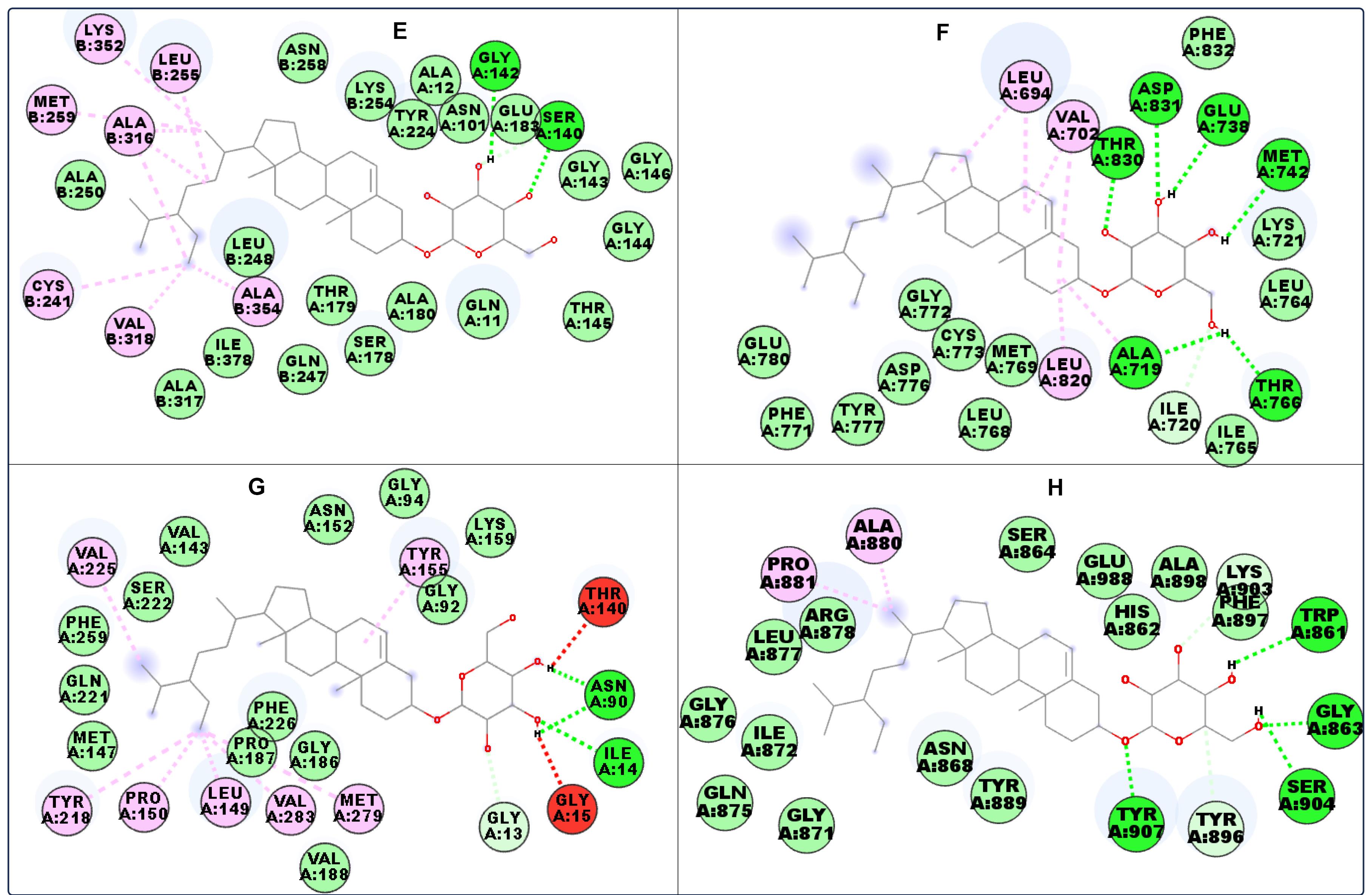
Figure 3.
Interacting residues of (E) Alpha-beta tubulin, (F) EGFR, (G) 17β-HSD1, and (H) PARP-1 with compound 1b. Green: Conventional hydrogen bonding. Lilac: Alkyl/pi-alkyl interaction. Blue: Carbon hydrogen bonding. Purple: Pi-sigma bonding. Light green: van der Waals forces
.
Interacting residues of (E) Alpha-beta tubulin, (F) EGFR, (G) 17β-HSD1, and (H) PARP-1 with compound 1b. Green: Conventional hydrogen bonding. Lilac: Alkyl/pi-alkyl interaction. Blue: Carbon hydrogen bonding. Purple: Pi-sigma bonding. Light green: van der Waals forces
17β-HSD1 converts weakly active oestrone to a pharmacologically more active estrogen, 17β-oestradiol, in the breast tissue using nicotinamide adenine dinucleotide (NADH) or its phosphate (NADPH) as a cofactor. 17β-oestradiol is crucial for the growth of breast cancer cells.56 The two compounds interacted with important 17β-HSD1 residues involved in its catalytic functions, which include tyrosine (TYR) 155 and lysine (LYS) 159 (Figures 2C and 3G).57 EGFR residues interacted with the compounds via alkyl, van der Waals, and hydrogen bonds (Figures 2B and 3F). The sugar moiety was used to interact with the protein via hydrogen bonding. In PARP-1, compound 1a formed a π-sigma bond with the protein in addition to alkyl, π-alkyl, van der Waals, and hydrogen bondings (Figures 2D and 3H). Since most of the selected proteins are expressed in the tested cancer cell lines, it can be suggested that the in silico study supports the in vitro study.
Drug-likeness study
In 1997, Christopher Lipinski from Pfizer introduced the “Rule of 5.” It suggests that orally administered drugs are less likely to be absorbed or permeate well if they have a molecular weight exceeding 500 g/mol, more than 5 hydrogen bond donors, more than 10 hydrogen bond acceptors, or a CLog P greater than 5.58 Although the “Rule of Five” is not sacrosanct because many blockbuster drugs in the clinic fail it, it has greatly helped in lead optimisation and drug design. In drug development, a drug candidate is assessed for its absorption, distribution, metabolism and elimination.35 Table 4 shows the physicochemical parameters and ADMET descriptors for the isolated compounds 1a and 1b as virtually estimated by the SwissADME software. The compounds have molecular weights greater than 500 g/mol, which violate the Rule of 5. However, other parameters (Log P, hydrogen bond acceptor and hydrogen bond donor) are within acceptable ranges. The compounds are also not inhibitors of metabolic enzymes.
Table 4.
Physicochemical/ADME descriptors of the isolated compounds
|
Parameters
|
1a
|
1b
|
| Molecular weight (g/mol) |
574.83 |
576.85 |
| Num. rotatable bonds |
8 |
9 |
| Num. H-bond acceptors |
6 |
6 |
| Num. H-bond donors |
4 |
4 |
| TPSA (Å2) |
99.38 |
99.38 |
| Log Po/w |
4.23 |
4.98 |
| CYP1A2 inhibitor |
No |
No |
| CYP2C19 inhibitor |
No |
No |
| CYP2C9 inhibitor |
No |
No |
| CYP2D6 inhibitor |
No |
No |
| CYP3A4 inhibitor |
No |
No |
| Synthetic accessibility |
7.93 |
8.02 |
The bioavailability radar helps in the initial evaluation of how drug-like a molecule is, using the following physicochemical properties: Lipophilicity (LIPO), Flexibility (FLEX), Insaturation (INSATU), Insolubility (INSOLU), Polarity (POLAR), and Size (SIZE) (Figure 4). Lipophilicity is measured by Log P, which ranges from −0.7 to + 5.0. Size is assessed by molecular weight ( ≤ 500 g/mol). Polarity is indicated by TPSA ( ≤ 140 Å2). Insolubility is represented by log S. Insaturation is evaluated based on the fraction of carbon atoms in sp³ hybridization. Flexibility is measured by the number of rotatable bonds ( ≤ 9).59 The bioavailability radar in Figure 4 shows that the bioavailability rating of the two compounds was slightly within the suitable physicochemical space for oral bioavailability (www.swissadme.ch).
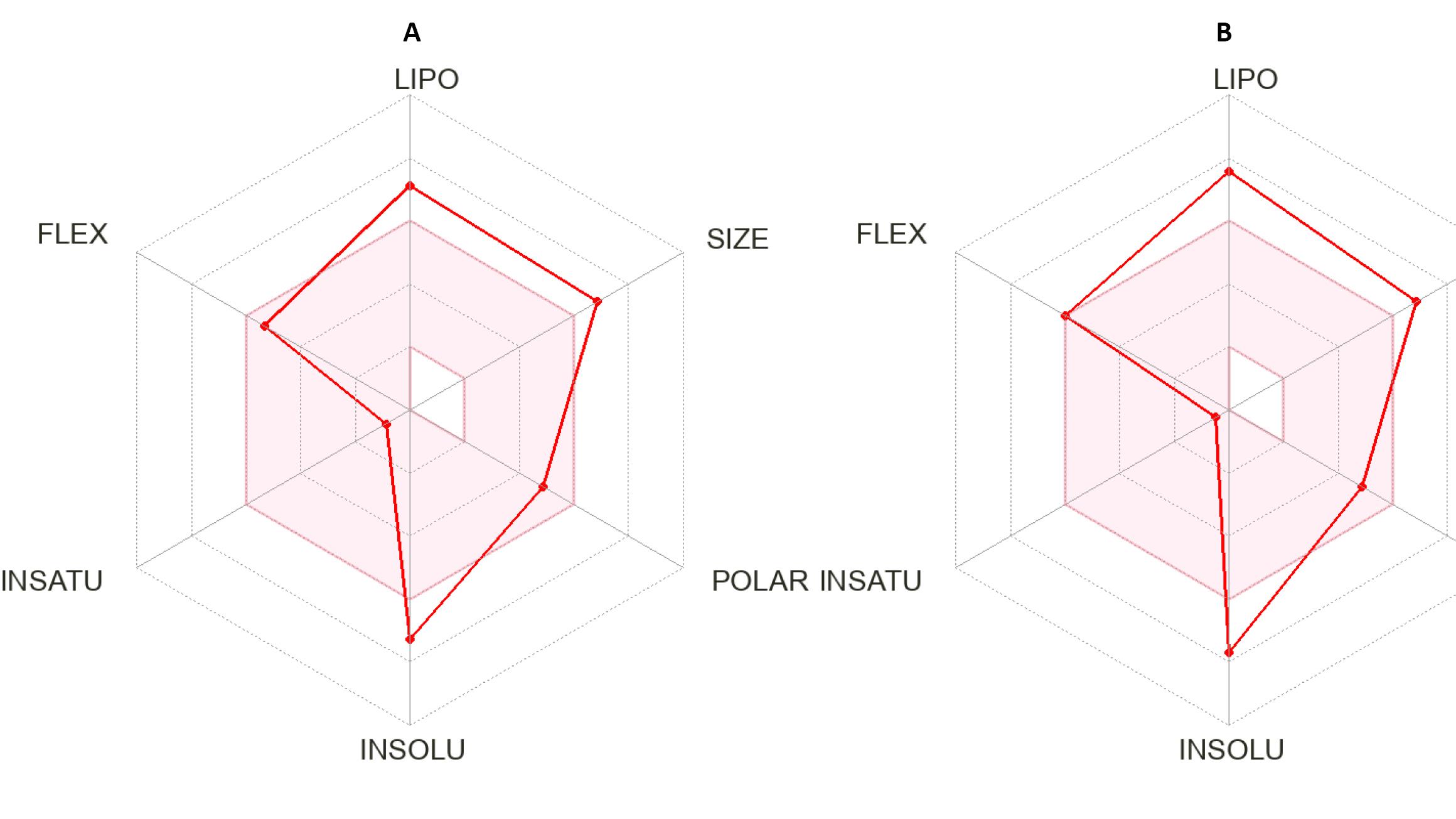
Figure 4.
Bioavailability radar for the isolated compounds 1a (A) and 1b (B). LIPO: Lipophilicity. FLEX: Flexibility. INSATU: Insaturation. INSOLU: Insolubility. POLAR: Polarity. SIZE: Size
.
Bioavailability radar for the isolated compounds 1a (A) and 1b (B). LIPO: Lipophilicity. FLEX: Flexibility. INSATU: Insaturation. INSOLU: Insolubility. POLAR: Polarity. SIZE: Size
Conclusion
Medicinal plants are a productive source of drugs, including anticancer agents. In this study, stigmast-5,22-dien-3-O-β-D-glucoside and sitosterol-3-O-β-D-glucoside were isolated from the methanol root extract of O. subscorpioidea. Their structures were established by various spectroscopic data analyses (FTIR (Figure S8), 1D & 2D NMR). The results of the MTT assay show that the compounds were cytotoxic against human cervical (HeLa) and breast (MCF-7) cancer cell lines, with IC50 values of 37.0 ± 4.51 and 137.07 ± 19.43 µg/mL, respectively. The results of the docking analysis of the isolated compounds showed they had strong binding affinities for the colchicine-binding site of alpha-beta tubulin, epidermal growth factor receptor kinase, poly (ADP-ribose) polymerse-1, and 17β-hydroxysteroid dehydrogenase-1. To the best of our knowledge, this is the first report of the isolation of these compounds from O. subscorpioidea. These isolated compounds may, therefore, contribute to the cytotoxic effects of O. subscorpioidea.
Competing Interests
The authors declare no conflicts of interests.
Ethical Approval
Not applicable.
Supplementary Files
Supplementary file 1 contains Table S1 and Figures S1-S8.
(pdf)
Acknowledgements
Yemi A. Adekunle acknowledges the award of a Commonwealth Split-Site PhD Scholarship by the Government of the United Kingdom to visit Liverpool John Moores University for part of his PhD research (NGCN-2021-184). Lutfun Nahar gratefully acknowledges the financial support of the European Regional Development Fund - Project ENOCH (No. CZ.02.1.01/0.0/0.0/16_019/0000868) and the Czech Agency Grants - Project 23-05474S and Project 23-05389S.
References
- Sarker SD, Nahar L, Miron A, Guo M. Anticancer natural products. In: Sarker SD, Nahar L, eds. Annual Reports in Medicinal Chemistry. Vol 55. Academic Press; 2020. p. 45-75. doi: 10.1016/bs.armc.2020.02.001.
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 2020; 83(3):770-803. doi: 10.1021/acs.jnatprod.9b01285 [Crossref] [ Google Scholar]
- Ahmad Khan MS, Ahmad I. Herbal medicine: current trends and future prospects. In: Ahmad Khan MS, Ahmad I, Chattopadhyay D, eds. New Look to Phytomedicine. Academic Press; 2019. p. 3-13. doi: 10.1016/b978-0-12-814619-4.00001-x.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71(3):209-49. doi: 10.3322/caac.21660 [Crossref] [ Google Scholar]
- Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A. Cancer statistics for the year 2020: an overview. Int J Cancer 2021; 149(4):778-89. doi: 10.1002/ijc.33588 [Crossref] [ Google Scholar]
- Kuete V, Efferth T. African flora has the potential to fight multidrug resistance of cancer. Biomed Res Int 2015; 2015:914813. doi: 10.1155/2015/914813 [Crossref] [ Google Scholar]
- Gyanani V, Haley JC, Goswami R. Challenges of current anticancer treatment approaches with focus on liposomal drug delivery systems. Pharmaceuticals (Basel) 2021; 14(9):835. doi: 10.3390/ph14090835 [Crossref] [ Google Scholar]
- Hashmi MA, Khan A, Hanif M, Farooq U, Perveen S. Traditional uses, phytochemistry, and pharmacology of Olea europaea (Olive). Evid Based Complement Alternat Med 2015; 2015:541591. doi: 10.1155/2015/541591 [Crossref] [ Google Scholar]
- Huang YL, Oppong MB, Guo Y, Wang LZ, Fang SM, Deng YR. The Oleaceae family: a source of secoiridoids with multiple biological activities. Fitoterapia 2019; 136:104155. doi: 10.1016/j.fitote.2019.04.010 [Crossref] [ Google Scholar]
- Mansour KA, Elbermawi A, Al-Karmalawy AA, Lahloub MF, El-Neketi M. Cytotoxic effects of extracts obtained from plants of the Oleaceae family: bio-guided isolation and molecular docking of new secoiridoids from Jasminum humile. Pharm Biol 2022; 60(1):1374-83. doi: 10.1080/13880209.2022.2098346 [Crossref] [ Google Scholar]
- Odoma S, Umar Zezi A, Mohammed Danjuma N, Ahmed A, Garba Magaji M. Elucidation of the possible mechanism of analgesic actions of butanol leaf fraction of Olaxsubscorpioidea Oliv. J Ethnopharmacol 2017; 199:323-7. doi: 10.1016/j.jep.2016.12.052 [Crossref] [ Google Scholar]
- Agbabiaka TO, Adebayo IA. The medicinal properties of Olaxsubscorpioidea. In: Bhat RA, Hakeem KR, Dervash MA, eds. Phytomedicine. Academic Press; 2021. p. 555-80. doi: 10.1016/b978-0-12-824109-7.00019-4.
- Adekunle Y, Samuel BB, Fatokun AA, Nahar L, Sarker SD. Olaxsubscorpioidea Oliv (Olacaceae): an ethnomedicinal and pharmacological review. J Nat Prod Discov 2022; 1(2):673. doi: 10.24377/jnpd.article673 [Crossref] [ Google Scholar]
- Soladoye MO, Amusa NA, Raji-Esan SO, Chukwuma EC, Taiwo AA. Ethnobotanical survey of anti-cancer plants in Ogun state, Nigeria. Ann Biol Res 2010; 1(4):261-73. [ Google Scholar]
- Gbadamosi IT, Erinoso SM. A review of twenty ethnobotanicals used in the management of breast cancer in Abeokuta, Ogun state, Nigeria. Afr J Pharm Pharmacol 2016; 10(27):546-64. doi: 10.5897/ajpp2015.4327 [Crossref] [ Google Scholar]
- Segun PA, Ogbole OO, Ajaiyeoba EO. Medicinal plants used in the management of cancer among the Ijebus of southwestern Nigeria. J Herb Med 2018; 14:68-75. doi: 10.1016/j.hermed.2018.04.002 [Crossref] [ Google Scholar]
- Adekunle YA, Samuel BB, Nahar L, Fatokun AA, Sarker SD. Cytotoxic triterpenoid saponins from the root of Olaxsubscorpioidea Oliv (Olacaceae). Phytochemistry 2023; 215:113853. doi: 10.1016/j.phytochem.2023.113853 [Crossref] [ Google Scholar]
- Adekunle Y, Samuel B, Emmanuel T, Adeniji J, Ogbole O. Cytotoxicity analysis of nineteen medicinal plants extracts on breast adenocarcinoma (MCF-7) and rhabdomyosarcoma (RD) cancer cell lines. Trends Pharm Sci 2022; 8(1):57-64. doi: 10.30476/tips.2022.94313.1136 [Crossref] [ Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65(1-2):55-63. doi: 10.1016/0022-1759(83)90303-4 [Crossref] [ Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004; 25(13):1605-12. doi: 10.1002/jcc.20084 [Crossref] [ Google Scholar]
- Adewumi AT, Oluyemi WM, Adekunle YA, Adewumi N, Alahmdi MI, Soliman ME. Propitious indazole compounds as β‐ketoacyl‐ACP synthase inhibitors and mechanisms unfolded for TB cure: integrated rational design and MD simulations. ChemistrySelect 2023; 8(3):e202203877. doi: 10.1002/slct.202203877 [Crossref] [ Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H. The protein data bank. Nucleic Acids Res 2000; 28(1):235-42. doi: 10.1093/nar/28.1.235 [Crossref] [ Google Scholar]
- Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 35 Å resolution. J Mol Biol 2001; 313(5):1045-57. doi: 10.1006/jmbi.2001.5077 [Crossref] [ Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004; 428(6979):198-202. doi: 10.1038/nature02393 [Crossref] [ Google Scholar]
- Lee D, Long SA, Adams JL, Chan G, Vaidya KS, Francis TA. Potent and selective nonpeptide inhibitors of caspases 3 and 7 inhibit apoptosis and maintain cell functionality. J Biol Chem 2000; 275(21):16007-14. doi: 10.1074/jbc.275.21.16007 [Crossref] [ Google Scholar]
- Mazza C, Breton R, Housset D, Fontecilla-Camps JC. Unusual charge stabilization of NADP + in 17beta-hydroxysteroid dehydrogenase. J Biol Chem 1998; 273(14):8145-52. doi: 10.1074/jbc.273.14.8145 [Crossref] [ Google Scholar]
- Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell 2000; 6(4):909-19. doi: 10.1016/s1097-2765(05)00089-4 [Crossref] [ Google Scholar]
- Dawicki-McKenna JM, Langelier MF, DeNizio JE, Riccio AA, Cao CD, Karch KR. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol Cell 2015; 60(5):755-68. doi: 10.1016/j.molcel.2015.10.013 [Crossref] [ Google Scholar]
- Li Y, Zhao J, Gutgesell LM, Shen Z, Ratia K, Dye K. Novel pyrrolopyridone bromodomain and extra-terminal motif (BET) inhibitors effective in endocrine-resistant ER + breast cancer with acquired resistance to fulvestrant and palbociclib. J Med Chem 2020; 63(13):7186-210. doi: 10.1021/acs.jmedchem.0c00456 [Crossref] [ Google Scholar]
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem 2002; 277(48):46265-72. doi: 10.1074/jbc.M207135200 [Crossref] [ Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 1998; 95(7):927-37. doi: 10.1016/s0092-8674(00)81717-1 [Crossref] [ Google Scholar]
- Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J. BI 6727, a polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res 2009; 15(9):3094-102. doi: 10.1158/1078-0432.Ccr-08-2445 [Crossref] [ Google Scholar]
- Oluyemi WM, Samuel BB, Adewumi AT, Adekunle YA, Soliman MES, Krenn L. An allosteric inhibitory potential of triterpenes from Combretum racemosum on the structural and functional dynamics of Plasmodium falciparum lactate dehydrogenase binding landscape. Chem Biodivers 2022; 19(2):e202100646. doi: 10.1002/cbdv.202100646 [Crossref] [ Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010; 31(2):455-61. doi: 10.1002/jcc.21334 [Crossref] [ Google Scholar]
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017; 7:42717. doi: 10.1038/srep42717 [Crossref] [ Google Scholar]
- Kamal N, Clements C, Gray AI, Edrada-Ebel R. Anti-infective activities of secondary metabolites from Vitex pinnata. J Appl Pharm Sci 2016; 6(1):102-6. doi: 10.7324/japs.2016.600117 [Crossref] [ Google Scholar]
- Falodun A, Ali S, Quadir IM, Choudhary IM. Phytochemical and biological investigation of chloroform and ethylacetate fractions of Euphorbia heterophylla leaf (Euphorbiaceae). J Med Plants Res 2008; 2(12):365-9. [ Google Scholar]
- Khatun M, Billah M, Quader MA. Sterols and sterol glucoside from Phyllanthus species. Dhaka Univ J Sci 2012; 60(1):5-10. doi: 10.3329/dujs.v60i1.10327 [Crossref] [ Google Scholar]
- Ridhay A, Noor A, Soekamto NH, Harlim T, van Altena I. A stigmasterol glycoside from the root wood of Melochiaumbellata (Houtt) Stapf var degrabrata K. Indones J Chem 2012; 12(1):100-3. doi: 10.22146/ijc.21379 [Crossref] [ Google Scholar]
- Rosli N, Khairuddean M, Jamain Z. Phytochemical and biological activity studies of the leaves of Garcinia hombroniana Pierre. Malays J Chem 2020; 22(4):81-102. [ Google Scholar]
- Peshin T, Kar HK. Isolation and characterization of β-sitosterol-3-O-β-D-glucoside from the extract of the flowers of Viola odorata. Br J Pharm Res 2017; 16(4):1-8. doi: 10.9734/bjpr/2017/33160 [Crossref] [ Google Scholar]
- Quradha MM, Khan R, Adhikari A, Rauf A, Rashid U, Bawazeer S. Isolation, biological evaluation, and molecular docking studies of compounds from Sophora mollis (Royle) Graham ex Baker. ACS Omega 2021; 6(24):15911-9. doi: 10.1021/acsomega.1c01532 [Crossref] [ Google Scholar]
- Ekhuemelo DO, Agbidye FS, Anyam JV, Ekhuemelo C, Igoli JO. Antibacterial activity of triterpenes from the stem bark and heartwood of Erythrophleumsuaveolens (Guill & Perr) Brenan. J Appl Sci Environ Manag 2019; 23(5):783-9. doi: 10.4314/jasem.v23i5.2 [Crossref] [ Google Scholar]
- Erwin Erwin, Pusparohmana WR, Safitry RD, Marliana E, Usman Usman, Kusuma IW. Isolation and characterization of stigmasterol and β-sitosterol from wood bark extract of Baccaurea macrocarpa Miq Mull Arg. Rasayan J Chem 2020; 13(4):2552-8. doi: 10.31788/rjc.2020.1345652 [Crossref] [ Google Scholar]
- Khan ME, Odokpe AU, Tor-Anyiin TA. Isolation and characterization of stigmasterol and β-sitosterol from Cassia sieberiana (Fabaceae) leaf extract. J Chem Soc Nigeria 2020; 45(1):135-42. [ Google Scholar]
- Shwe HH, Thein WW, Win SS, Pe NN, Win T. Structural characterization of stigmasterol and β-sitosterol from the roots of Premnaherbacea Roxb. Int Eur Ext Enablement Sci Eng Manag 2019; 7(8):195-201. [ Google Scholar]
- Masoko P, Nemudzivhadi V. Isolation and chemical structural characterization of the mixture of two related phytosterols from Ricinus communis L (Euphorbiaceae) leaves. European J Med Plants 2015; 10(2):1-11. doi: 10.9734/ejmp/2015/18172 [Crossref] [ Google Scholar]
- Elfita E, Muharni M, Latief M, Darwati D, Widiyantoro A, Supriyatna S. Antiplasmodial and other constituents from four Indonesian Garcinia spp. Phytochemistry 2009; 70(7):907-12. doi: 10.1016/j.phytochem.2009.04.024 [Crossref] [ Google Scholar]
- Ikpefan EO, Ayinde BA, Omeje EO, Azhar M, Farooq AD, Shah ZA. Isolation and anti-cancer evaluation of two anti-proliferative constituents from the chloroform fraction of leaves of Conyza sumatrensis(Retz) E H Walker, Asteraceae. Sci Afr 2021; 13:e00854. doi: 10.1016/j.sciaf.2021.e00854 [Crossref] [ Google Scholar]
- Hamza RA, Mostafa I, Mohamed YS, Dora GA, Ateya AM, Abdelaal M. Bioguided isolation of potential antitumor agents from the aerial parts of cultivated cardoon (Cynara cardunculus var altilis). Saudi Pharm J 2023; 31(1):125-34. doi: 10.1016/j.jsps.2022.11.011 [Crossref] [ Google Scholar]
- Zeng J, Liu X, Li X, Zheng Y, Liu B, Xiao Y. Daucosterol inhibits the proliferation, migration, and invasion of hepatocellular carcinoma cells via Wnt/β-catenin signaling. Molecules 2017; 22(6):862. doi: 10.3390/molecules22060862 [Crossref] [ Google Scholar]
- Vo TK, Ta QTH, Chu QT, Nguyen TT, Vo VG. Anti-hepatocellular-cancer activity exerted by β-sitosterol and β-sitosterol-glucoside from Indigofera zollingeriana Miq. Molecules 2020;25(13). doi: 10.3390/molecules25133021.
- Hou X, Du J, Zhang J, Du L, Fang H, Li M. How to improve docking accuracy of AutoDock42: a case study using different electrostatic potentials. J Chem Inf Model 2013; 53(1):188-200. doi: 10.1021/ci300417y [Crossref] [ Google Scholar]
- Horgan MJ, Zell L, Siewert B, Stuppner H, Schuster D, Temml V. Identification of novel β-tubulin inhibitors using a combined in silico/in vitro approach. J Chem Inf Model 2023; 63(20):6396-411. doi: 10.1021/acs.jcim.3c00939 [Crossref] [ Google Scholar]
- Naaz F, Haider MR, Shafi S, Yar MS. Anti-tubulin agents of natural origin: targeting taxol, vinca, and colchicine binding domains. Eur J Med Chem 2019; 171:310-31. doi: 10.1016/j.ejmech.2019.03.025 [Crossref] [ Google Scholar]
- Lawrence HR, Vicker N, Allan GM, Smith A, Mahon MF, Tutill HJ. Novel and potent 17beta-hydroxysteroid dehydrogenase type 1 inhibitors. J Med Chem 2005; 48(8):2759-62. doi: 10.1021/jm049045r [Crossref] [ Google Scholar]
- Frotscher M, Ziegler E, Marchais-Oberwinkler S, Kruchten P, Neugebauer A, Fetzer L. Design, synthesis, and biological evaluation of (hydroxyphenyl)naphthalene and -quinoline derivatives: potent and selective nonsteroidal inhibitors of 17beta-hydroxysteroid dehydrogenase type 1 (17beta-HSD1) for the treatment of estrogen-dependent diseases. J Med Chem 2008; 51(7):2158-69. doi: 10.1021/jm701447v [Crossref] [ Google Scholar]
- Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 2000; 44(1):235-49. doi: 10.1016/s1056-8719(00)00107-6 [Crossref] [ Google Scholar]
- Rai M, Singh AV, Paudel N, Kanase A, Falletta E, Kerkar P. Herbal concoction unveiled: a computational analysis of phytochemicals’ pharmacokinetic and toxicological profiles using novel approach methodologies (NAMs). Curr Res Toxicol 2023; 5:100118. doi: 10.1016/j.crtox.2023.100118 [Crossref] [ Google Scholar]